29 August 2008
WetLab
Midipreps
- Digested Midipreps of Biobricks - The midipreps of the following constructs were digested with EcoRI and SpeI in order to confirm the expected length of the biobrick. The following biobricks were midi preped, digested and analysed on agarose gel electrophoresis:
- Terminator, size of biobrick = 129bp
- AK3, size of biobrick = Should be no insert present
- The gel below shows the results of this analysis,
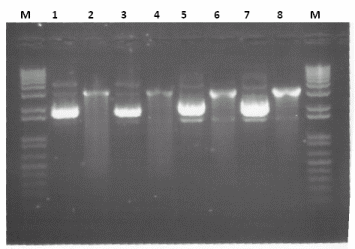 Lanes : M = Marker, 1 = Terminator Midi 1 Undigested, 2 = Terminator Midi 1 Digested, 3 = Terminator Midi 2 Undigested, 4 = Terminator Midi 2 Digested, 5 = AK3 Midi 1 Undigested, 6 = AK3 Midi 1 Digested, 7 = AK3 Midi 2 Undigested, 8 = AK3 Midi 2 Digested, PCR
In a pursuit to verify the various PCR protocols and PCR primers we carried out the following reactions:
- PCR with the pDR110 Vector using AmyE Fw1 and R1 primers
- PCR with B.subtilis genomic DNA using AmyE Fw1 and R1 primers
- Single Colony PCR using AmyE Fw1 and R1 primers and AmyE Fw1 and R2 primers
For each of these reactions a gradient of temperatures was used during the 10 cycles in the annealing stage of PCR and also the following 20 cycles, see our protocol for more information in summary the following reactions were carried out:
- 1. Vector - 10 cycles at 57oC, 20 cycles at 64oC, Primers = Fw1/R1
- 2. Vector - 10 cycles at 55oC, 20 cycles at 62oC, Primers = Fw1/R1
- 3. Vector - 10 cycles at 53oC, 20 cycles at 60oC, Primers = Fw1/R1
- 4. Genomic - 10 cycles at 57oC, 20 cycles at 64oC, Primers = Fw1/R1
- 5. Genomic - 10 cycles at 55oC, 20 cycles at 62oC, Primers = Fw1/R1
- 6. Genomic - 10 cycles at 53oC, 20 cycles at 60oC, Primers = Fw1/R1
- 7. Single Colony PCR - 10 cycles at 52oC, 20 cycles at 64oC, Primers = Fw1/R1, Sample of colony 1ul
- 8. Single Colony PCR - 10 cycles at 50oC, 20 cycles at 64oC, Primers = Fw1/R1, Sample of colony 1ul
- 9. Single Colony PCR - 10 cycles at 48oC, 20 cycles at 64oC, Primers = Fw1/R1, Sample of colony 1ul
- 10. Single Colony PCR - 10 cycles at 52oC, 20 cycles at 64oC, Primers = Fw1/R1,Sample of colony 2ul
- 11. Single Colony PCR - 10 cycles at 50oC, 20 cycles at 64oC, Primers = Fw1/R1,Sample of colony 2ul
- 12. Single Colony PCR - 10 cycles at 48oC, 20 cycles at 64oC, Primers = Fw1/R1, Sample of colony 2ul
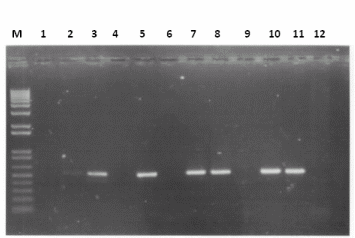
Results
- Vector PCR - From the results the optimum conditions is clearly reaction 3 (10 cycles at 53oC, 20 cycles at 60oC, Primers = Fw1/R1)
- Genomic - From the results the optimum conditions is clearly reaction 5 (10 cycles at 55oC, 20 cycles at 62oC, Primers = Fw1/R1)
- Single Colony PCR - From the results clearly it is sufficient to use only 1ul of sample. In addition only the combination of Fw1 and Rv1 is working at either 10 cycles at 52oC, 20 cycles at 64oC or 10 cycles at 50oC, 20 cycles at 64oC
|
 "
"