3 September 2008
Wetlab
Cloning
- Results of PCR reactions run on the 2nd analysed on a gel (results below - top gel)
- To check that PCR with Pfu was possible, a PCR reaction with the strongest vector PCR with taq was performed with Pfu under standard Pfu conditions[[1]] but a longer annealing time (1 minute); the primers used were AmyE 5' forward and reverse primers at 52°C for the first 10 cycles then 60°C for 20 cycles. Aad9 PCR was again tested according to the standard protocol and agin at 52°C for the first 10 cycles then 58°C or 56°C for 20 cycles (results below - lower gel)
- GFP-Terminator biobrick and RFP-Terminator biobrick cut again with XbaI and SpeI to produce biobrick vectors for our PCR clones
- GFP-Terminator biobrick and RFP-Terminator biobrick cut again with EcoRI and SpeI to produce biobrick vectors for our GeneArt produced clones
- Biobrick digest run on a gel for gel purification of vector
- GeneArt constructs in vectors midiprepped from Xl1-Blue cells
Transformation of B.subtilis
- Previously we have tried to test efficient integration into the B.subtilis genome. However, the primers we were using have so far failed to yield either a positive or negative result when a signle colony PCR was carried out on transformed B.subtilis. In order to check firstly if the primers are working and secondly if the single colony PCR protocol is working we carried out a series of testing. First to test the verification primers we are going to carry out a PCR on purified genome, that we have previously used successfully in a PCR reaction. We set up the following conditions:
- 1 cycle 95oC for 30 seconds
- 30 cycles, 30 seconds 95oC, 1 minute at 60/58/56oC, 5 minutes at 72oC
- A negative control was used at 56oC where no genomic DNA was added.The results are shown in figure 1.1 below.
- To confirm if the single colony PCR protocol is working we used this protocol with a new set of primers and a set that has been primers shown to work with purified genomic DNA. AmyE fw1 and rv1 (shown to work) are primers used to purify the 5' integration site from B.subtilis and AmyE fw2 and rv2 (not tested yet) are primers used to purify the 3'integration sites from B.subtilis. We performed the single colony PCR with Fw1 and Rv2 (not tested) and Fw1 and Rv1 (tested) under the following conditions:
- Using the primer, Fw1 and Rv2 -
- 1 cycle 95oC for 30 seconds,
- 10 cycles, 30 seconds 95oC, 1 minute at 52/50/48oC, 5 minutes at 72oC
- 20 cycles, 30 seconds 95oC, 1 minute at 61oC, 5 minutes at 72oC
- 1 cycle, 1 minute at 72oC.
- Using the primers, Fw1 and Rv1 -
- 1 cycle 95oC for 30 seconds,
- 10 cycles, 30 seconds 95oC, 1 minute at 48oC, 5 minutes at 72oC
- 20 cycles, 30 seconds 95oC, 1 minute at 61oC, 5 minutes at 72oC
- 1 cycle, 1 minute at 72oC.
- A negative control was also used where no primers were added.
Results
 A 1% Agarose gel showing the results of various PCR reactions Upper Lanes:
- Lane 1- Pfu PCR reaction, first 10 cycles at 52oC and then 20 cycles at 60oC using AmyE 5' integration sequence primers, (effectively a positive control)
- Lane 2- Pfu PCR reaction, first 10 cycles at 50oC and then 20 cycles at 65oC using AmyE 3' integration sequence primers,
- Lane 3- Pfu PCR reaction, first 10 cycles at 48oC and then 20 cycles at 65oC using AmyE 3' integration sequence primers,
- Lane 4- Pfu PCR reaction, first 10 cycles at 56oC and then 20 cycles at 65oC using LacI gene primers,
- Lane 5- Taq PCR reaction, first 10 cycles at 56oC and then 20 cycles at 65oC using LacI gene primers but no template DNA (negative control)
- Lane 6- Taq PCR reaction, first 10 cycles at 54oC and then 20 cycles at 60oC using Aad9 primers,
- Lane 7- Taq PCR reaction, first 10 cycles at 52oC and then 20 cycles at 60oC using Aad9 primers,
- Lane 8- Taq PCR reaction, first 10 cycles at 50oC and then 20 cycles at 60oC using Aad9 primers,
- Lane 9 - Taq PCR reaction, first 10 cycles at 50oC and then 20 cycles at 60oC using Aad9 primers but no template DNA
- As can be observed all the PCRs failed, including the positive control. The most likely explanation is that the reactions were left to long before PCR started (in the case of Pfu) or that a vital reaction component was not correctly added to the reactions. A trial of Pfu will need to be carried out to determine if the enzyme itself may be a problem
Lower Lanes:
- Lane 1 - Annealing step 60oC using verification primers,
- Lane 2- Annealing step 58oC using verification primers,
- Lane 3- Annealing step 56oC using verification primers,
- Lane 4- Annealing step 56oC using verification primers, no DNA negative control,
- Lane 5 - Annealing step 52oC using AmyE fw1 and rev2 primers,
- Lane 6- Annealing step 50oC using AmyE fw1 and rev2 primers,
- Lane 7- Annealing step 48oC using AmyE fw1 and rev2 primers,
- Lane 8- Annealing step 61oC using AmyE fw1 and rev1, positive control,
- Lane 9 - Annealing step 48oC using AmyE fw1 and rev2, no DNA negative control,
- As we can see only for the positive control we see a band, this means that the verfication primers and the fw1 and rev 2 combinations are not working. We will double check the primer sequences and carry on trying to optimise the protocol.
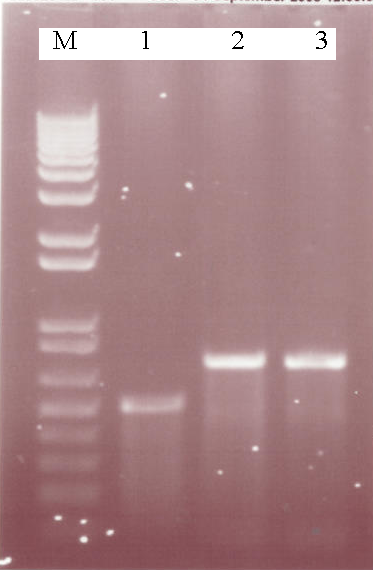 A 1% Agarose gel showing the results of various PCR reactions, M = Marker, Lane 1 - AmyE 5' Integration Sequence, Lane 2 - Aad9 PCR (58 oC), Lane 3 - Aad9 PCR (56 oC) - The AmyE 5' integration sequence was succesfully produced by Pfu. This DNA can now be cut and ligated into a biobrick for use in our constructs. Aad9 was succesfully produced using Taq, providing conditions for use with Pfu for biobrick cloning
Dry Lab
Motility
- Manual tracking of synthetic data was continued. Error bars were plotted and uploaded onto the OWW Wiki.
|