TUDelft/4 September 2008
From 2008.igem.org
| July | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | 6 | |
| 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| 21 | 22 | 23 | 24 | 25 | 26 | 27 |
| 28 | 29 | 30 | 31 | |||
| August | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | ||||
| 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| 11 | 12 | 13 | 14 | 15 | 16 | 17 |
| 18 | 19 | 20 | 21 | 22 | 23 | 24 |
| 25 | 26 | 27 | 28 | 29 | 30 | 31 |
| September | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 8 | 9 | 10 | 11 | 12 | 13 | 14 |
| 15 | 16 | 17 | 18 | 19 | 20 | 21 |
| 22 | 23 | 24 | 25 | 26 | 27 | 28 |
| 29 | 30 | |||||
| October | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | ||
| 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| 27 | 28 | 29 | 30 | 31 | ||
Contents |
September 4th
Transformation
3A Assembly
Though unknown whether ligation worked, we tried to transform cells with total volume (15 ul) of our ligation. Plated on Chloroamphenicol o/n @ 37ºC.
PCR
A new PCR with the Pfx enzyme was performed on the E.coli genes. This time more template DNA was used as the last times this PCR was performed it yielded no results. Two reactions of 50 ul were set up as last time. 50ng of template (genomic E. coli DNA) was used, other characteristics were the same.
Strangely, only one of the reactions yielded a result, for the idi gene. The picture of the gel with band cut out can be seen above. At this point, it is not clear what went wrong.
Double restriction
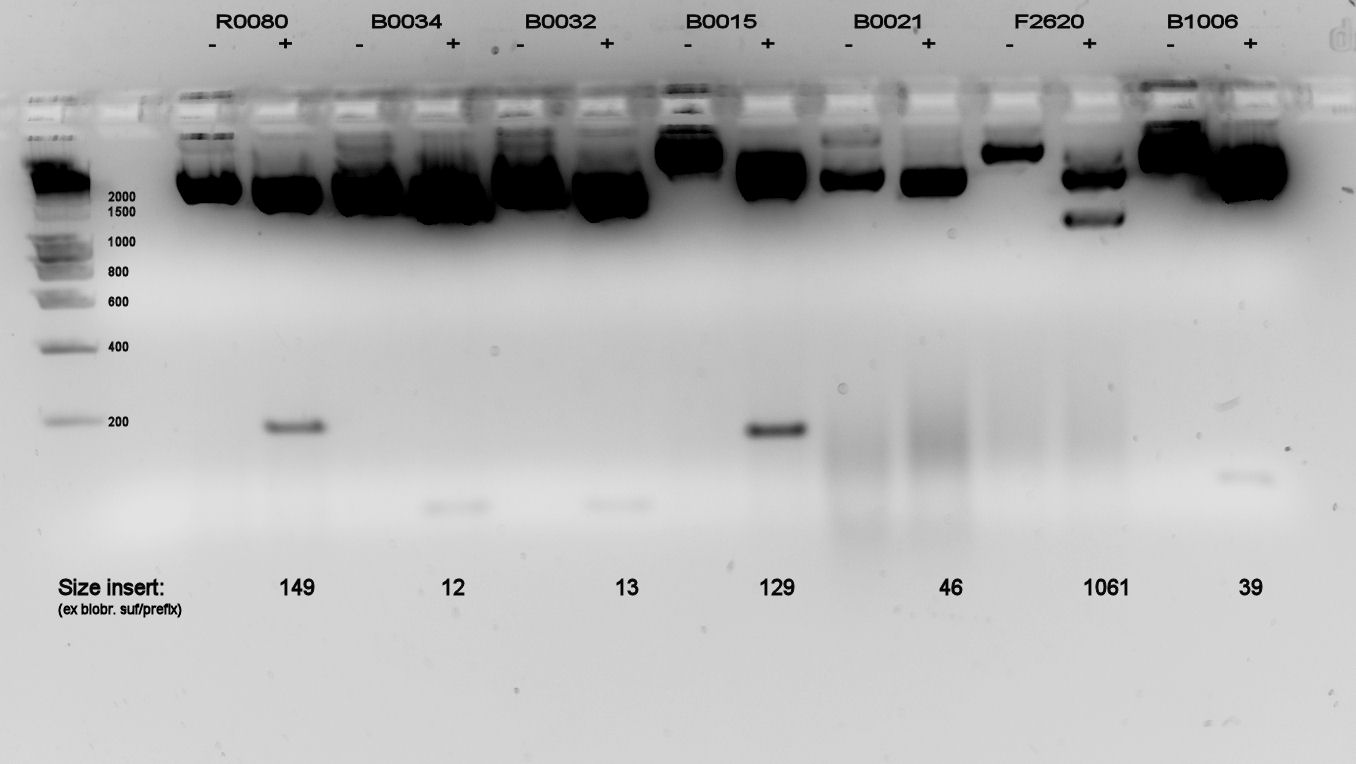
We did a double restriction (EcoRI & PstI) on the newly received living stabs, and ran a 2% agarose gel, because there were many parts < 200 bp. The gel is displayed in figure 1.
Most parts look all right. Due to the small parts having low mass and thus low light emittance, we used a lot of DNA, causing overexposure for the backbones. Only parts B0021 and F2620, a terminator and promoter, showed a smear in both cut and uncut DNA. B0021 was indeterminable, F2620 was correct size (which was also visible on the gel of the 2nd of September which was not posted). All the other parts were also likely to be correct size.
 "
"