|
Home
The Team
Project Report
Parts
Modeling
Notebook
Safety
CoLABoration
|
_3D-modeling
DNA-Origami
Planning the input pattern on the DNA-Origami surface, we generated various 3D-models of the huge molecule using NanoEngineer, Pymol and SwissPdbViewer (both freeware). Some of those models are shown here to give you an impression of the molecules we have engineered this year.
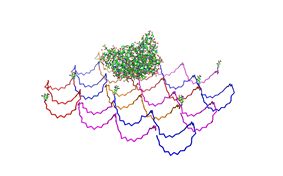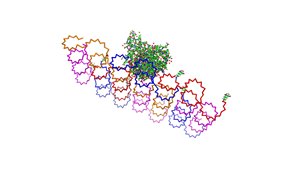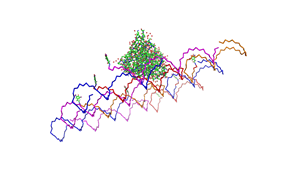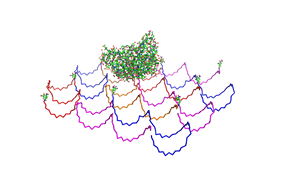 Fig.1: Some of the oligonucleotides that shape the single-stranded template fused to NIP-molecules and an anti-NIP-Fv-Fragment |
|
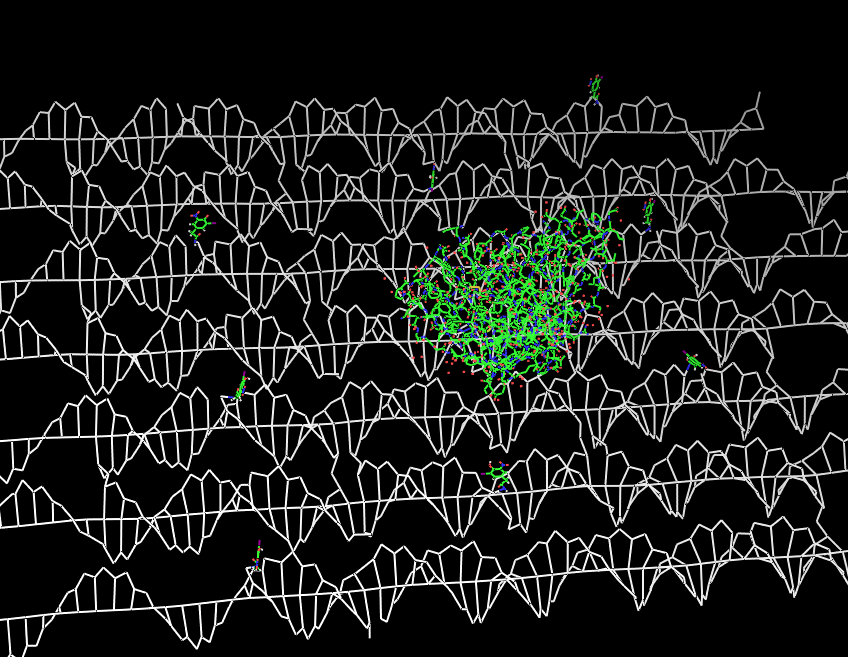 Fig.2: DNA-Origami with NIP-molecules and anti-NIP-fragment |
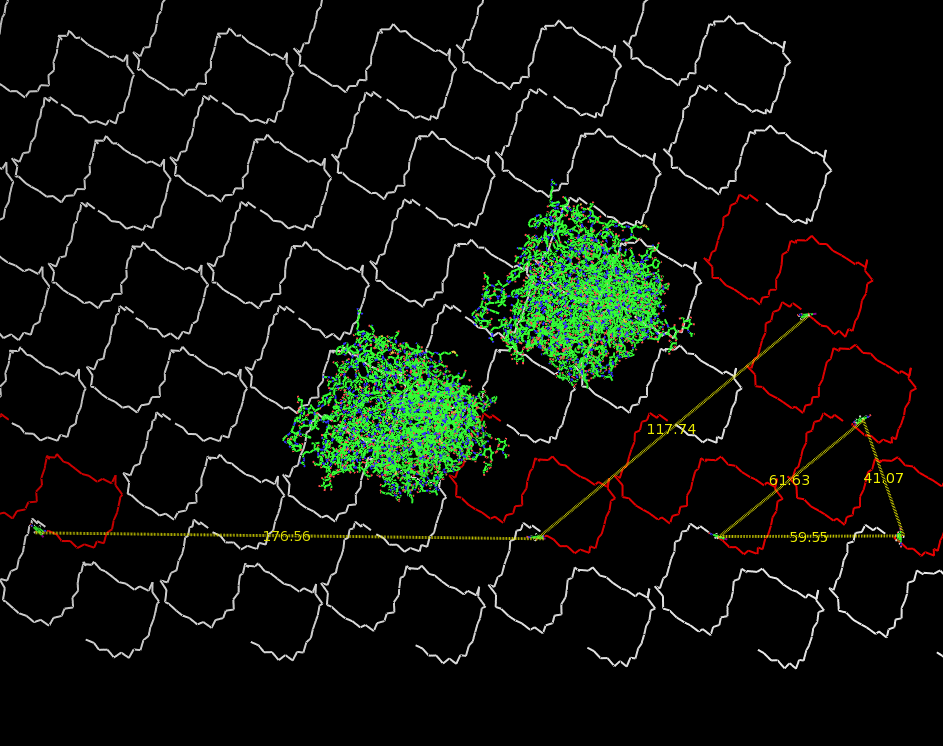 Fig.3: Oligo-pattern with scale and two fragments (distance estimation model, distances given in Ångstrom) |
 Fig.4: Oligo-pattern with scale and two T-Cell-receptors (distance estimation model, distances given in Ångstrom) |
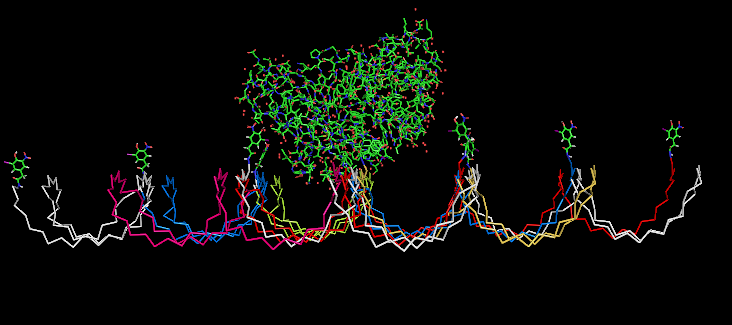 Fig.5: Oligo-pattern with NIP-molecules and Fab-fragment, binding accessibility estimation model
|
Fusion Proteins
The pictures below show models of the fusion proteins of our composite parts Bba_K157032 and Bba_K157033 code for (generated with "SwissPdbViewer"):
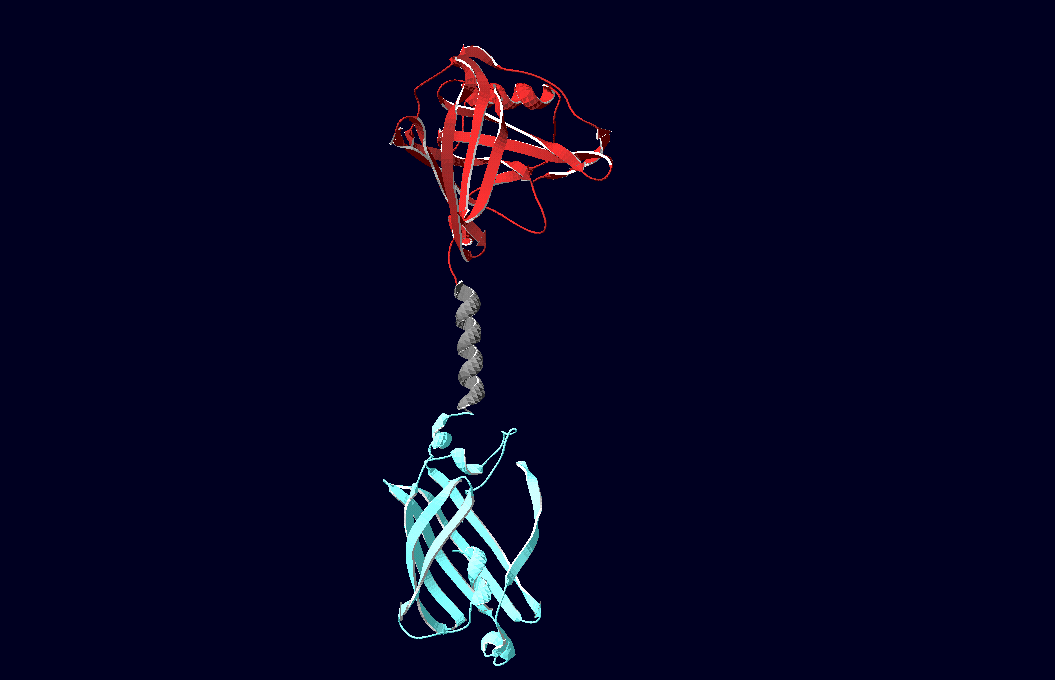 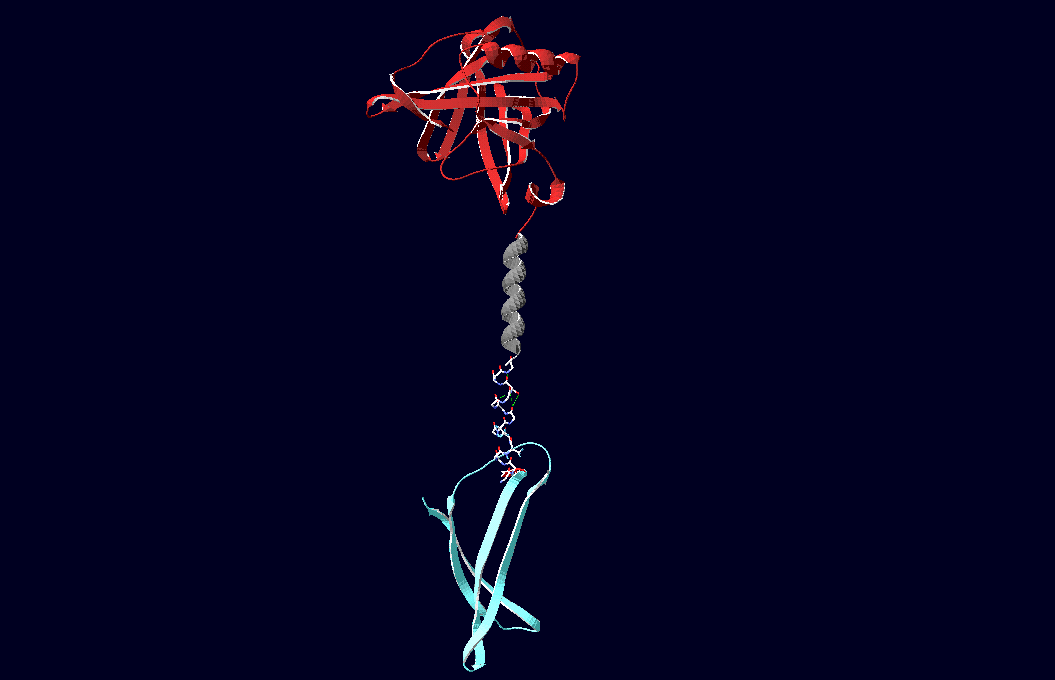
Fig.6: Red: LipocalinFluA, Grey: "Transmembrane region (BCR, no structural data; symbolized by helix), Blue: left: Split-Cerulean (CFP), N-terminal fragment; right: Split-Cerulean (CFP), C-terminal fragment
Following pdb-files have been used to generate the models:
Cerulean-CFP: 2Q57
Venus-YFP: 1MYW
Engineered Lipocalin: 1n0s
B1-8 Fv-Fragment:1a6w
|
 "
"







