Team:Hawaii/Construction of BioBrick intermediates
From 2008.igem.org
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Contents |
Construction of BioBrick intermediates
- Construct:
- nir + rbs
- GFP + tt
- GFPf + tt
Methods
Attempt #1
Restriction digest
- Digested nir, GFP, and GFPf with EcoRI and SpeI in NEBuffer 2
- Digested B0034 (rbs) and B0024 (tt) with EcoRI and XbaI in NEBuffer 2 with BSA
Gel purification
- Ran digest reaction on an EtBr stained 2% gel to extract bands of interest
Ligated parts
- Ligated parts in 20 mu;l reactions:
- 4 μl B0034 (8.8 ng) + 2.42 μl nir (26.378 ng)
- 1 μl B0024 (15.5. ng) + 8 μl GFP (41.6 ng)
- 0.4 μl B0024 (6.2 ng) + 1.8 μl GFPf (1.8 ng)
Subcloned in DH5α
- Used 5 μl ligation reaction to transform
- Incubated at 37C with shaking for 100 min.
Colony PCR verification of constructs
Attempt #2
Restriction digest
- Digested nir, GFP, and GFPf with EcoRI and SpeI in NEBuffer EcoRI with BSA
- Sequentially digested B0034 (rbs) and B0024 (tt) with XbaI and EcoRI in their respective buffers
Gel purification
- Ran digest reaction on an EtBr stained 1.2% gel to extract bands of interest
Ligated parts
- Ligated parts in 20 mu;l reactions:
Subcloned in DB3.1
- Used 5 μl ligation reaction to transform
Results
Attempt #1
Transformation
Colony forming units observed
| Construct | CFUs |
|---|---|
| nir+B0034 | 01 |
| GFP + B0024 | 5 |
| GFPf + B0024 | 15 |
1 Though no colonies were initially observed, when the plate was scraped and restreaked (to purify), colonies were observed on the new LB+amp plate.
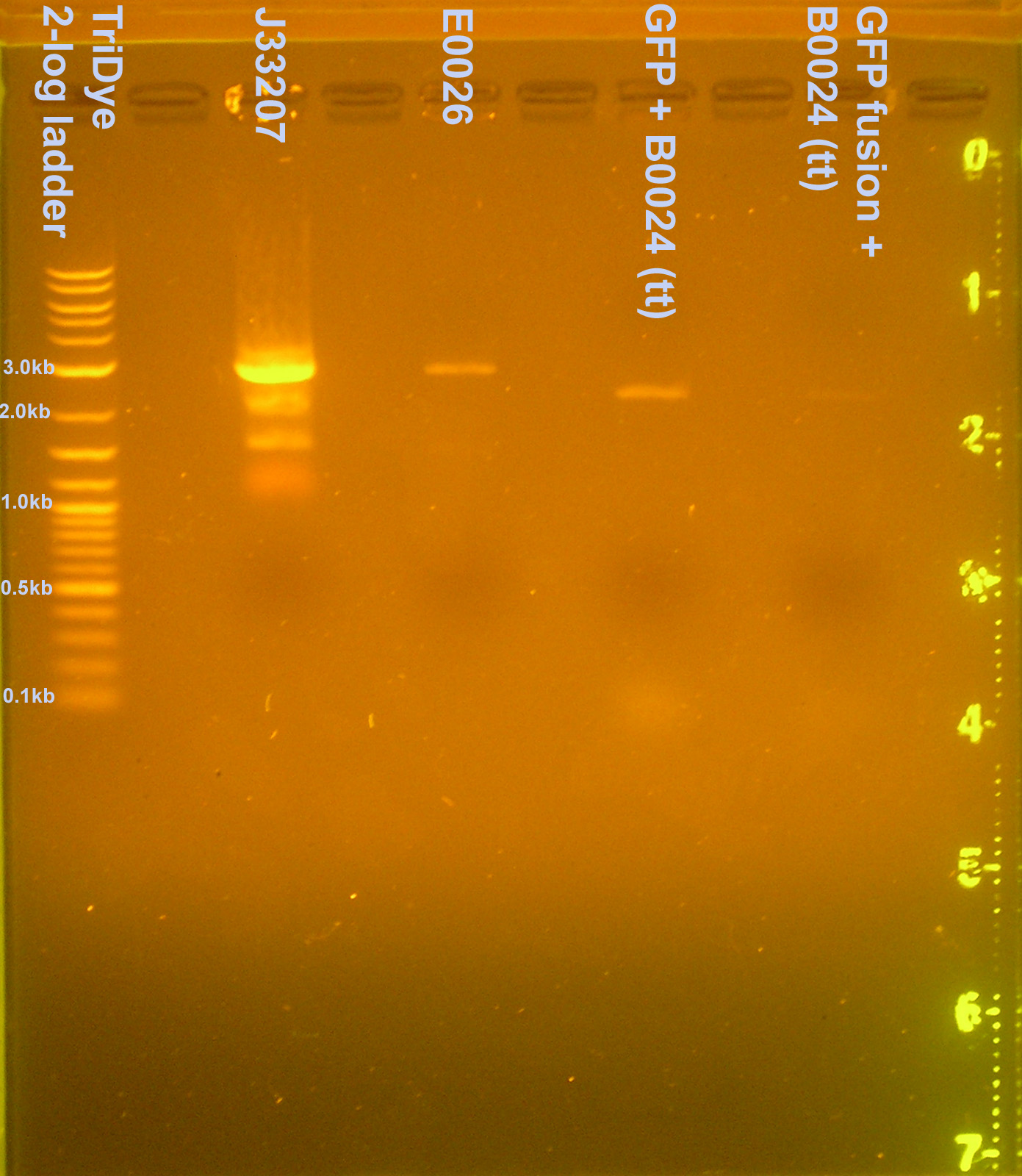
Restriction digest of plasmid preps (to verify insert)
The colony PCR was loaded onto a 4% gel so bands did not resolve. Since a plasmid prep was already done for the parts, the plasmid preps were RE digested with EcoRI to verify insertion of GFP(f) (Did not have room to load nir). The bands, expected to be ~2900bp, were both ~2100bp, indicating the GFP was not inserted. We likely digested with the wrong enzymes (XbaI and SpeI instead of EcoRI and XbaI) so the vector self ligated without the GFP(f) or tt present.
Discussion
- What was learned and how to do future experiments differently.
 "
"