Team:Hawaii/Notebook/2008-08- 6
From 2008.igem.org
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
Checked transformants from yesterday
- Grace
| Construct | Colony forming units |
|---|---|
| I14032 (plac) + B0030 (rbs) | 3 |
| pnir + B003 (rbs) | 0 |
| E0040 (GFP) + B0015 (tt) | 1 |
| GFPf + B0015 (tt) | 2 + 2 clusters of colonies |
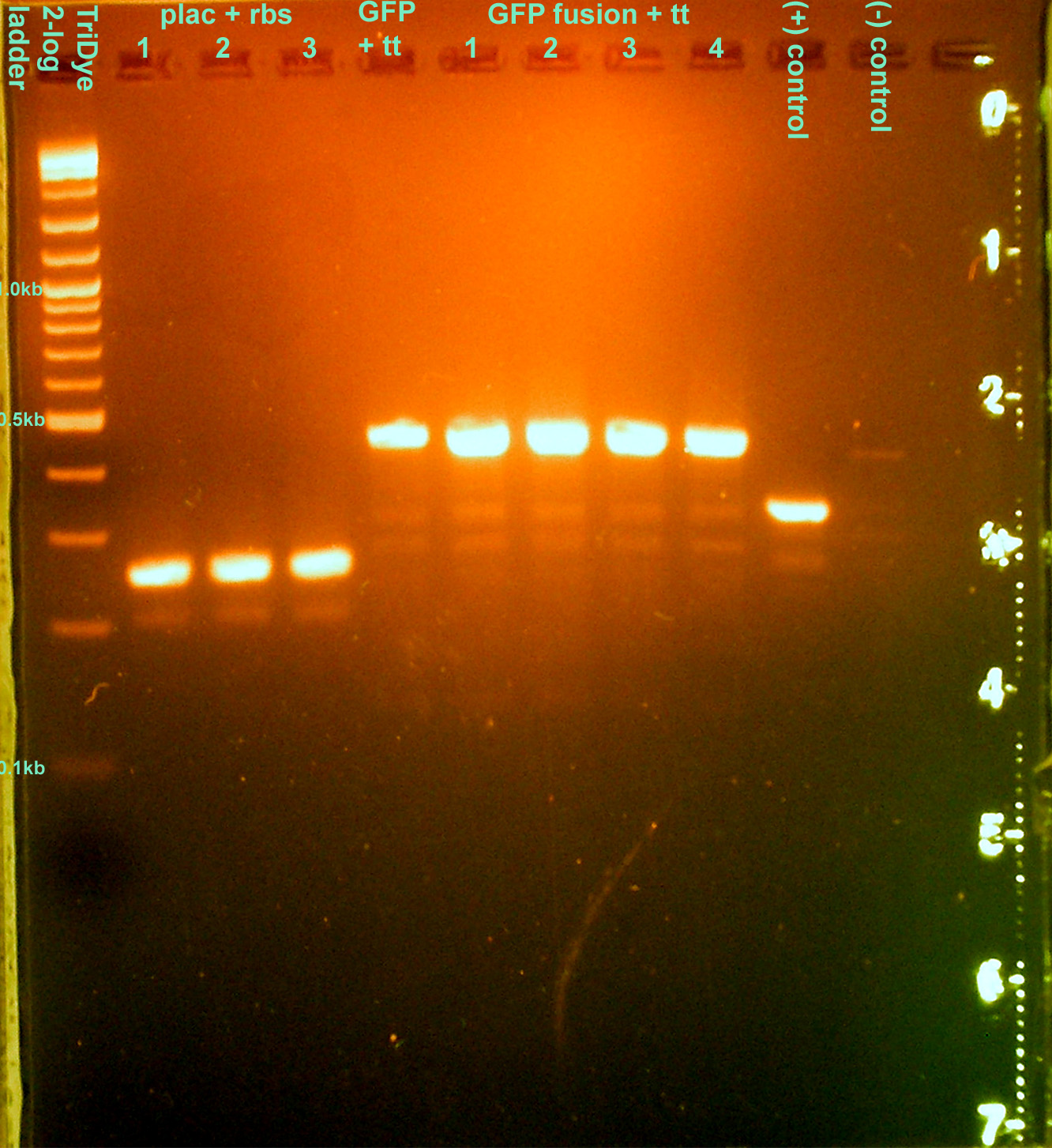
- Colony PCR'd transformants
- 30 cycles, anneal at 62C, extend for 1 min.
- Ran on EtBr stained 2.0% agarose gel at 95V for 1 hour
- None of the transformations were successful :o(
Determined DNA concentrations of purified RE'd PCR products from 8/4
- Grace
| DNA sample | Concentration |
|---|---|
| nir | 17.0 ng/μl |
| slr1 | 31.3 ng/μl |
| slr2 | 11.8 ng/μl |
| pilA | 13.0 ng/μl |
| GFP | 12.0 ng/μl |
| GFPf | 12.2 ng/μl |
| E0240 | 8.6 ng/μl |
| I14032 | 16.8 ng/μl |
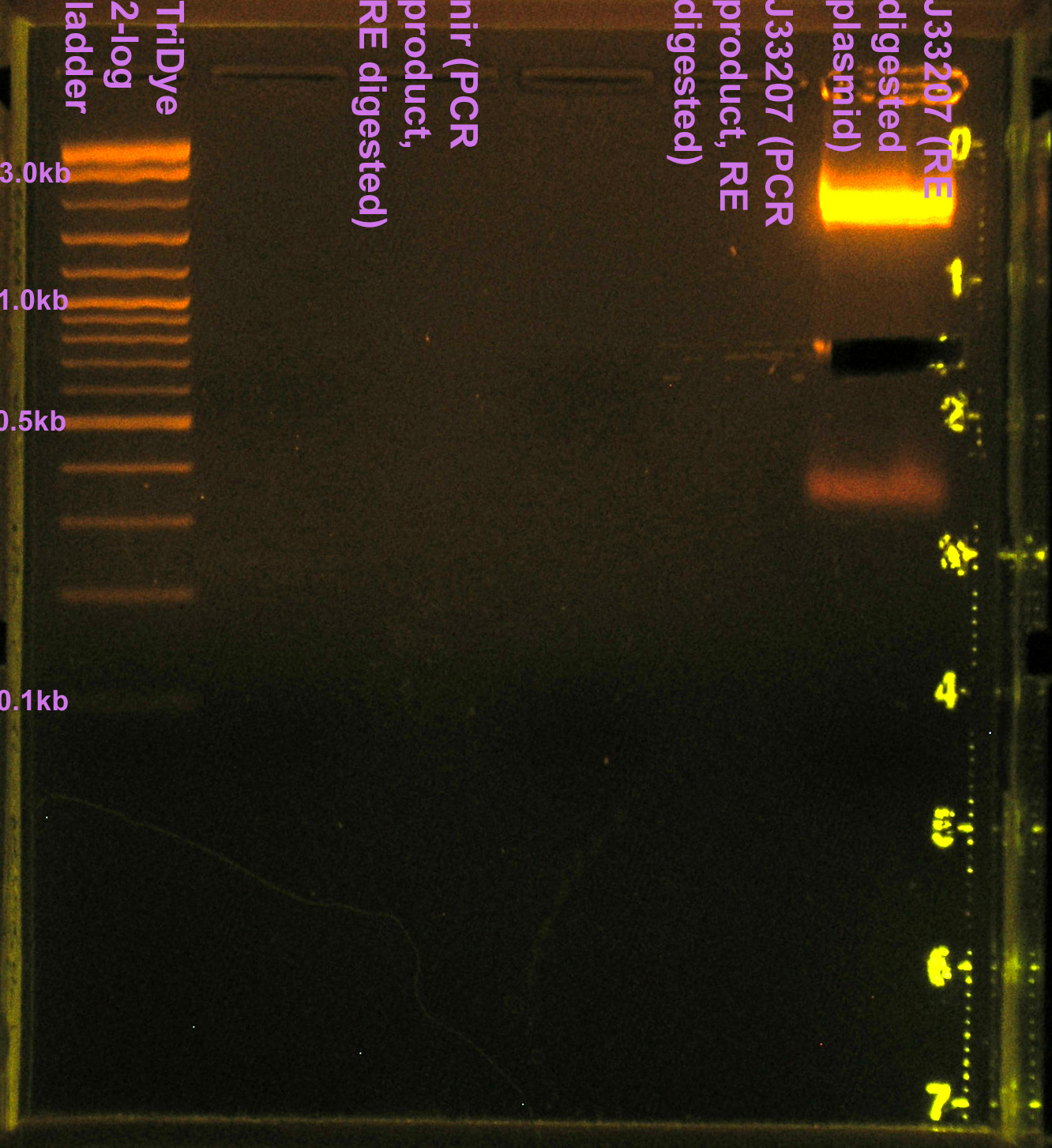
Prepared PCR'd nir and J33207 (from yesterday) for transformation/ligation
- Grace
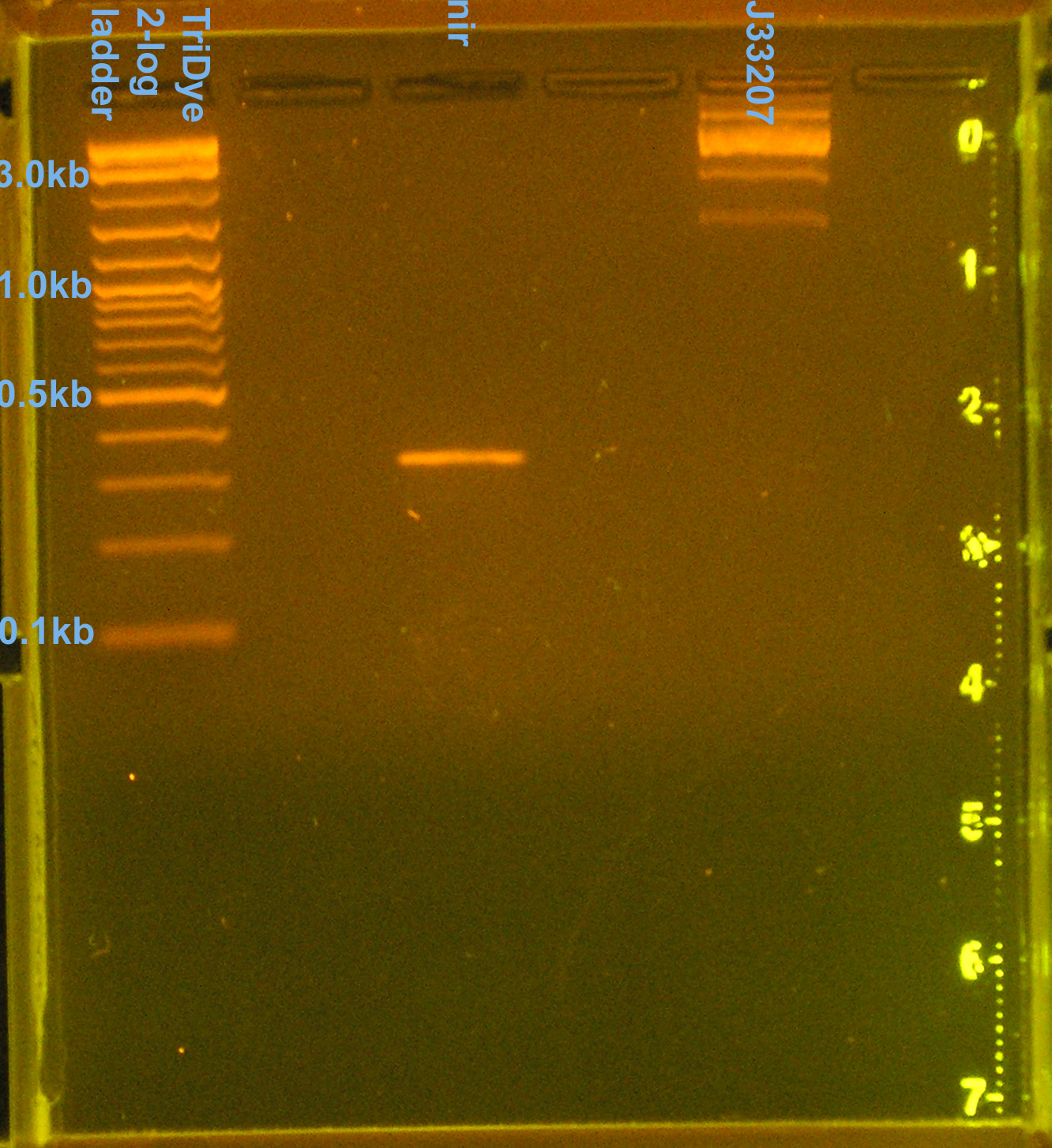
- Ran PCR products on EtBr stained 2.0% agarose gel at 60V for 100 min.
- nir band at 330bp confirmed
- J33207 = 4 bands (1.5kb, 2.5kb, 3.2kb, 10kb), none of which are correct
- partsregistry says J33207 DNA is inconsistent
- Extracted nir and 1.5kb J33207 band from gel
- RE digested in 50 μl rxns with EcoRI and SpeI in NEBuffer 2
- Larger rxn volume may improve digest efficiency
- Incubated at 37C for 2.5 hours
- RE digested 10 μl J33207 plasmid prep with EcoRI and SpeI in NEBuffer 2
- To confirm plasmid prep; if good, will use for ligation rxn
- Incubated at 37C for 2 hours
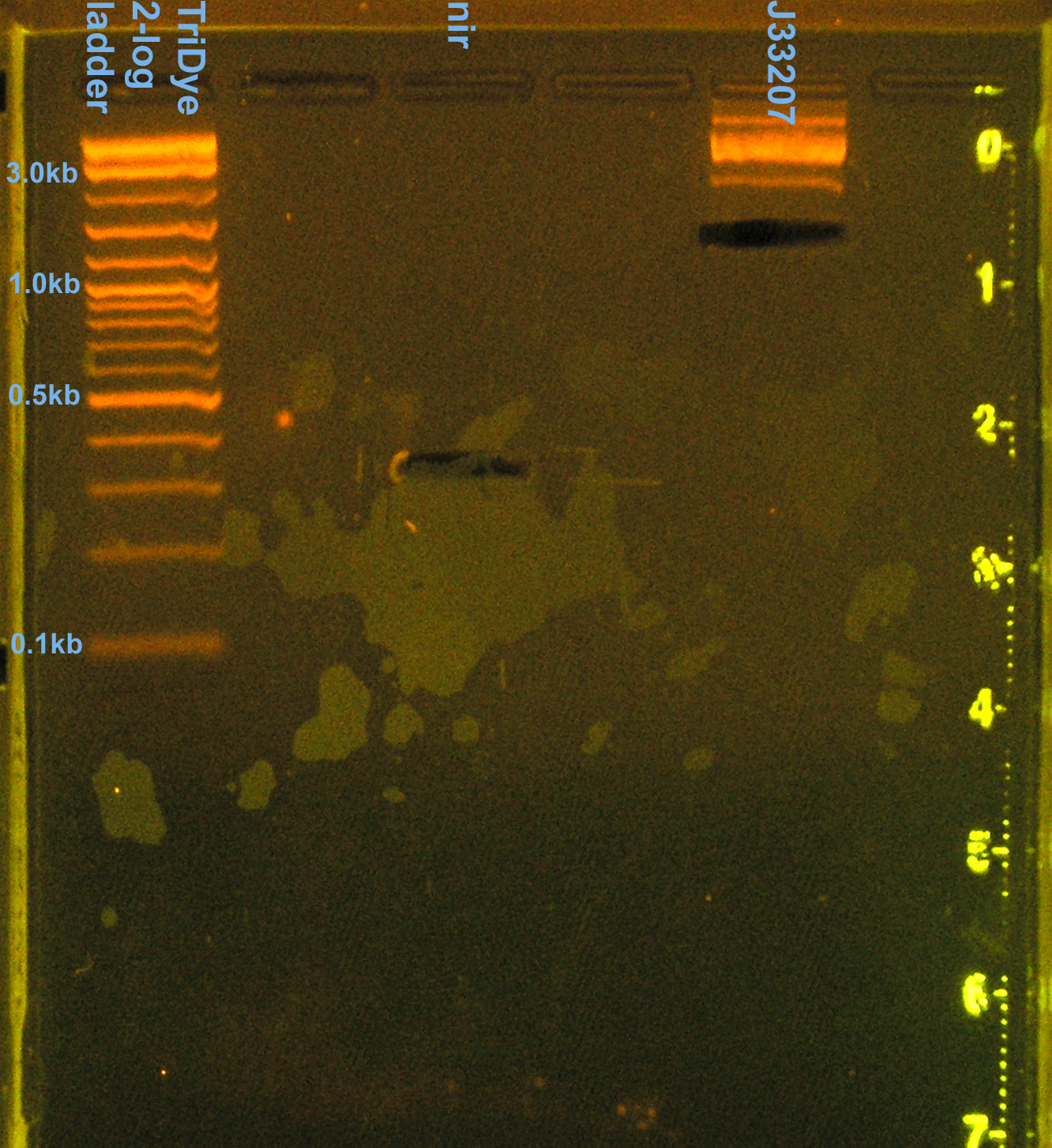
- Ran RE digests on EtBr stained 2.0% agarose gel at 72V for 90 min.
- No visible bands for nir and J33207 RE digest
- We lost a LOT of DNA in the gel purification.
- Band for J33207 plasmid prep RE digest should be ~650bp
- Band at ~850bp = VF2-VR = RE didn't cut???
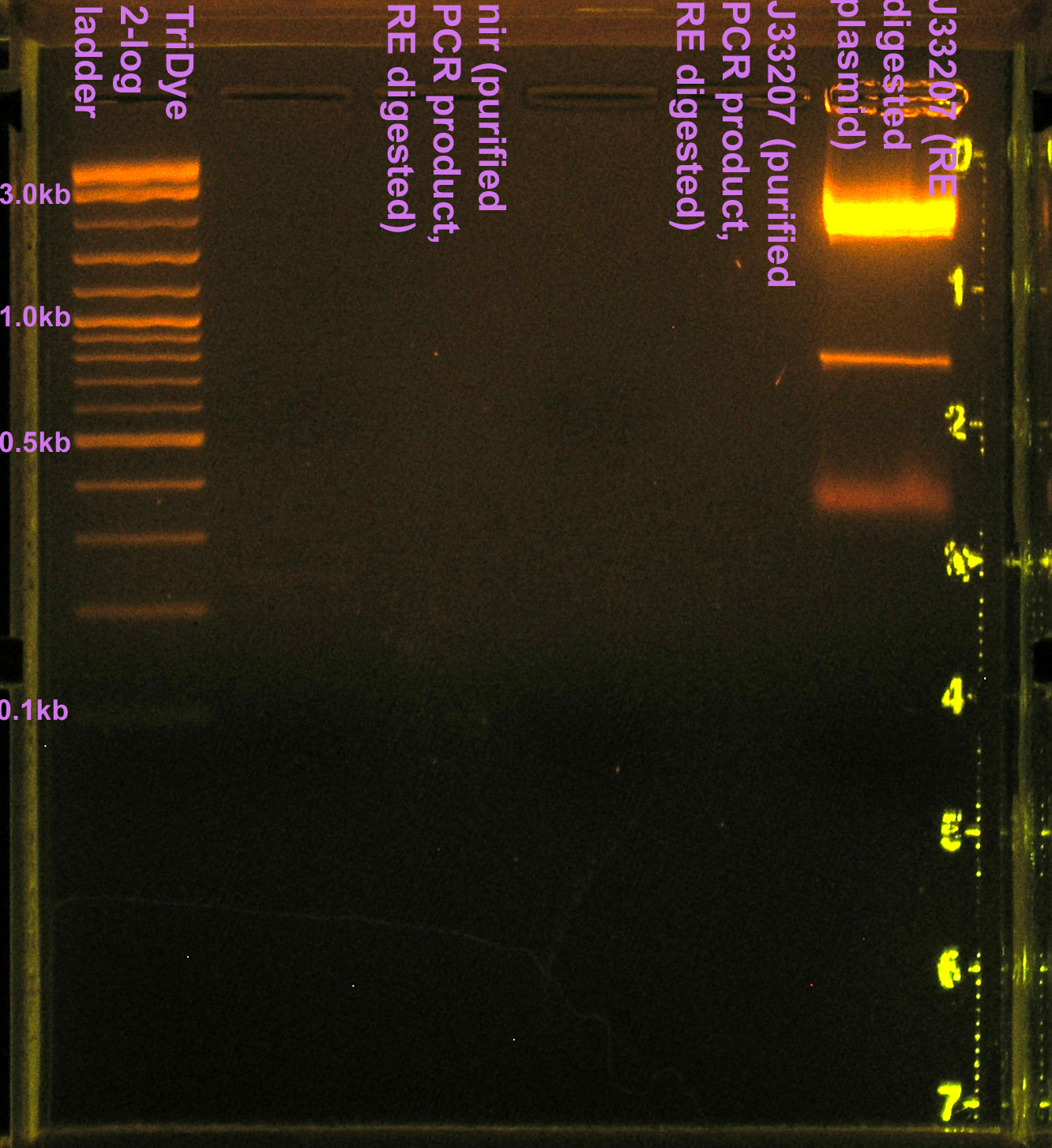
- Extracted J33207 band (~850) from gel
- Ligated 4.5 μl J33207 with 1.5 μl B0015 (tt)
- Krystle
- Ligated using quick ligase buffer:
- 3 μl gfpf with 1 μl B0015 (tt)
- 3 μl gfp with 1 μl B0015 (tt)
- 3 μl nir with 1 μl B0030 (rbs)
- 3 μl I14032 with 1 μl B0030 (rbs)
- Ligated using quick ligase buffer:
- Used 5 μl ligation reaction to transform 50 μl DB3.1 cells with the the 5 ligations mentioned above
Made LB+amp100 plates
- Grace and Margaret
Plasmid Prep
- Margaret
- pSMC121 was plasmid prepped today
- Krystle
- Started 200 ml preps of GFPf, nir, and pRL1383aM
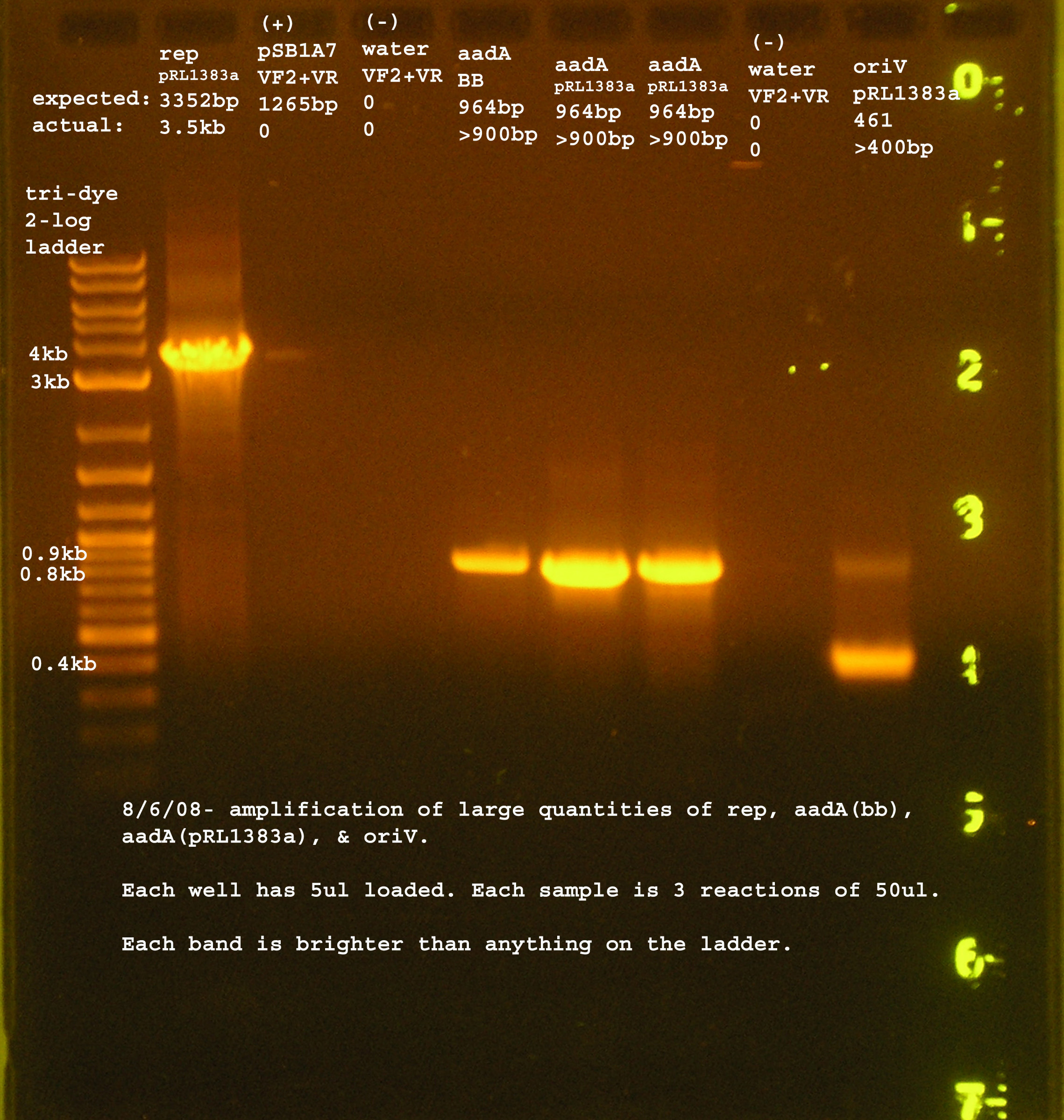
PCR
- Margaret
- made large quantities of aadA, rep, and oriV. Please refer to The experiment write-up for more details.
Drylab work
Sequencing
- Grace
- Reply from CORE Hawaii
- They will resequence slr1, slr2, BB-pRL1383a for free due to potential mix up of samples and poor quality reads
- For samples that were overloaded when 20ng/100bp sequencing guideline used, try 5 ng/100bp instead.
Discussion
- We're low on small nitrile gloves and Qiagen spin columns. Need to order more. NW
- Be careful when pouring agar plates. Bubbles suck!
 "
"