Team:Michigan/Project/Modeling/Model3
From 2008.igem.org
|
|---|
|
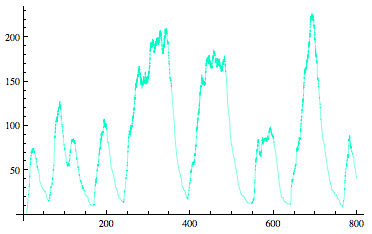
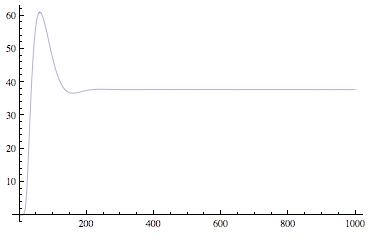
Sequestillator Model 3: Stochastic ModelUsing Mathematica's xCellerator program, we ran a Gillespie Integration of our system, and its corresponding ODE simulation with the same parameters (appropriately scaled to fit in the ODE context of concentration vs. stochastic context of number of proteins). All reactions were accounted for, including the hexamerization of NifA (which was defined by the binding of a dimer and tetramer of NifA. A tetramer of NifA arose from two dimers binding together). Stochastic Result-------------------------ODE Result
We could not run Ninfa index searches using the Indexilator on these stochastic simulations, due to the slowness of the xCellerator program and because our Indexilator can only evaluate ODE results, and not these noisy ones. |
|---|
 "
"