Team:Hawaii/Initial E. Coli Transformation
From 2008.igem.org
(Difference between revisions)
(attempt 4 added) |
(→Attempt 4) |
||
| Line 123: | Line 123: | ||
===Attempt 4=== | ===Attempt 4=== | ||
:*I had chosen the wrong plasmids, so I did another plasmid prep that is destined to win because of all my experience! | :*I had chosen the wrong plasmids, so I did another plasmid prep that is destined to win because of all my experience! | ||
| + | |||
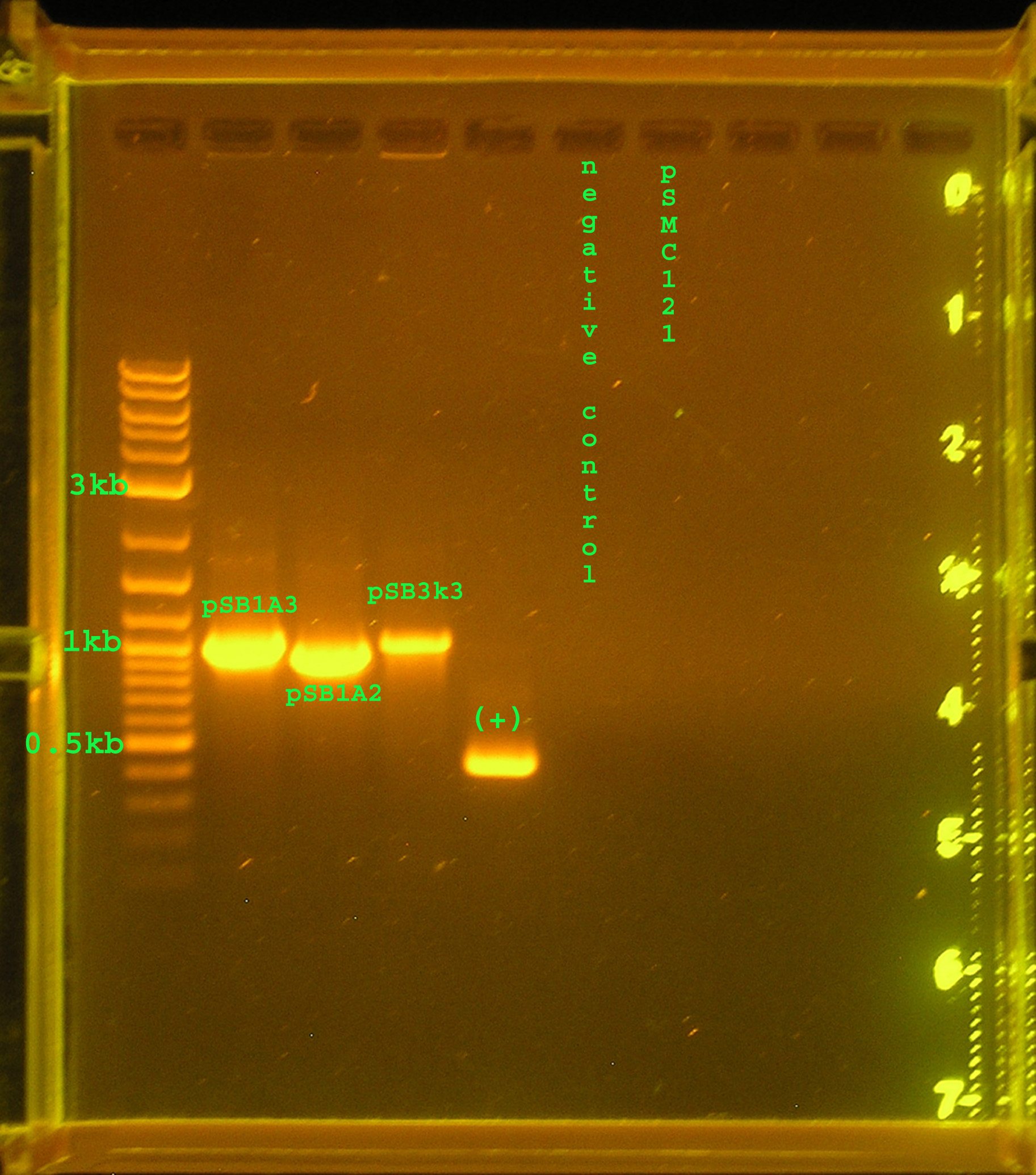
| + | [[Image:7_21_08_annotated.jpg|right|thumb|300px|Lane 1 contains the 2-log ladder, lane 2: pSB1A3's ccdB insert, lane3: pSB1A2's ccdB insert, lane 4: pSB3K3's ccdB insert, lane 5: positive control, lane 6: negative control, lane 7: pSMC121.]] | ||
{| border="1" | {| border="1" | ||
| Line 131: | Line 133: | ||
|- | |- | ||
! (+) pUC18 | ! (+) pUC18 | ||
| - | | | + | | lawn |
| - | | | + | |none |
|- | |- | ||
! (-) no plasmid | ! (-) no plasmid | ||
| - | | | + | | no colonies |
| - | | | + | | none |
|- | |- | ||
! pSB1A3 | ! pSB1A3 | ||
| - | | | + | | 1 colony |
| - | | | + | | restreaked, colony PCR |
|- | |- | ||
! pSB1A7 | ! pSB1A7 | ||
| - | | | + | | no colonies |
| - | | | + | |do-over |
|- | |- | ||
! BBa_I5102 | ! BBa_I5102 | ||
| - | | | + | | no colonies |
| - | | | + | | do-over |
|- | |- | ||
!BBa_J23012 | !BBa_J23012 | ||
| - | | | + | |no colonies |
| - | | | + | |do-over |
|} | |} | ||
Revision as of 04:04, 22 July 2008
Contents |
Transformation of DH5-alpha
Protocol by Invitrogen
Materials
Materials:
- 37°C shaking and non-shaking incubator
- 10 cm diameter LB agar plates with appropriate antibiotic (100 µg/ml ampicillin to select transformants containing pUC19 control DNA)
- LB, YT, or SOC Medium
- Dry ice and ethanol
- 37°C water bath
Before Starting
- Prepare a dry ice/ethanol bath and maintain at -70°C
- Equilibrate a water bath to 37°C
- Spread X-Gal onto LB agar plates with antibiotic, if desired
- Warm the medium and plates in a 37°C incubator for 30 minutes
- Obtain a test tube rack that will hold all transformation tubes so that they can all be put into a 37°C water bath at once.
Transformation Procedure
Note: Plasmid DNA should be free of phenol, ethanol, protein, and detergents for maximum transformation efficiency.
- Briefly centrifuge the ligation reaction and place on wet ice.
- Remove one 500 µl tube of DH5α™ cells and thaw on wet ice.
- Place the required number of sterile 1.5 ml microcentrifuge tubes on wet ice.
- Gently mix cells with the pipette tip and aliquot 50 or 100 µl into each microcentrifuge tube.
- Re-freeze any unused cells in the dry ice/ethanol bath for 5 minutes before returning the tube to the -70°C freezer.
- Pipet 1 to 5 µl (1-10 ng DNA) of each ligation reaction directly into the competent cells and mix by tapping gently. Do not mix by pipetting up and down. Store the remaining ligation reaction at -20°C.
- (Optional) Pipet 5 µl (500 pg) pUC19 control DNA into 100 µl competent cells and mix as described in Step 6.
- Incubate the vial on ice for 30 minutes.
- Heat-shock for exactly 20 seconds in the 37°C water bath for 50 µl volume (45 seconds for 100 µl transformation). Do not mix or shake.
- Remove vial from the 37°C bath and place on ice for 2 minutes.
- Add 900 to 950 µl of pre-warmed medium of choice to each vial. Proceed to next page.
- Place the vial in a microcentrifuge rack on its side and secure with tape to avoid loss of the vial. Shake the vial at 37°C for exactly 1 hour at 225 rpm in a shaking incubator.
- Spread 20 µl to 200 µl from each transformation vial on separate, labeled LB agar plates. We recommend that you plate two different volumes. You may have to dilute cells 1:10 to obtain well-spaced colonies. Generally ligations are at least 10-fold lower efficiency.
- (Optional) For cells transformed with pUC19 control DNA, plate 100 µl of the transformation reaction, then serially dilute the transformation reaction 1:10 and 1:100 and plate 100 µl of each dilution on plates containing 100 µg/ml ampicillin.
- Store the remaining transformation reaction at 4°C and plate out the next day, if desired. If necessary, cells may be concentrated by centrifuging in a microcentrifuge (5 seconds) and resuspending them in 100 µl. Plate 1 to 90 µl.
- Invert the plates and incubate at 37°C overnight.
Notes
- Transformation efficiency should be greater than 1 x 106 transformants/µg pUC19 DNA.
- To select transformants using blue/white screening, make sure that selective plates contain 50 µg/ml X-gal.
- Concentration of pUC19 provided in Invitrogen kit is 100 pg/µl.
Reference
"Subcloning Efficiency™ DH5α™ Competent Cells." 2006. Invitrogen. 01 June 2008. Available http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf.
Protocol from SC Lab
- Thaw cells on ice
- Add 10 µl ligation reaction with a pipetter and mix by flicking the tube gently
- Incubate the tube 30 minutes on ice
- Heat shock cells for 2 minutes at 42º C. To do this, transfer the cells directly from ice to a water bath at 42º C.
- Incubate on ice for 2 minutes
- Add 800 µl SOC media and transfer to a test tube
- Shake at 37º C for 2.5 hours (30 min is OK for Ampicillin resistance)
- Plate 200 µl of cells on selective media.
Notes
- Add 10 ml sterile 1M MgCl2 and 10 ml 1M MgSO4 to SOB media before use.
Questions
- Concentration of plasmid used?
- How much cells?
Protocol Used
- Thawed cells (2-17, OD600 = 0.3) on ice.
- Transformed 50 μl cells w/ 1 μl pRL1383a plasmid (10 pg per μl)
- Incubated on ice 30 min.
- Heat shocked 60 sec. in 42C water bath
- Added 250 μl SOC
- Incubated @ 37C for 1 hour with shaking (235 rpm)
- Plated 20 μl onto LB+spectinomycin plates
Results
80 colonies were observed 16 hours after innoculation. Transformation efficiency = 4 transformants/μl
Discussion
Transformation of DB3.1 Competent Cells
Attempts 1-3
| Plasmid | Description of Plate | Further Action |
|---|---|---|
| (+) pUC18 | lawn/lawn/lawn | none needed |
| (-) no plasmid | nothing/nothing/nothing | none needed |
| pSB3K3 | 1 colony | restreaked, plasmid prep |
| pSB1A2 | no colonies/none/2 colonies | watch plate for one more day/none/restreaked |
| pSB1AK3 | no colonies | watch plate for one more day/none/none |
Attempt 4
- I had chosen the wrong plasmids, so I did another plasmid prep that is destined to win because of all my experience!
| Plasmid | Description of Plate | Further Action |
|---|---|---|
| (+) pUC18 | lawn | none |
| (-) no plasmid | no colonies | none |
| pSB1A3 | 1 colony | restreaked, colony PCR |
| pSB1A7 | no colonies | do-over |
| BBa_I5102 | no colonies | do-over |
| BBa_J23012 | no colonies | do-over |
 "
"