Team:Bologna/UV radiation/
From 2008.igem.org
(→E.coli SOS System) |
(→E.coli SOS System) |
||
| Line 32: | Line 32: | ||
E.Coli cells have a system that recovery DNA damage when it occurs and it is the best studied transcriptional response ''[1]''. | E.Coli cells have a system that recovery DNA damage when it occurs and it is the best studied transcriptional response ''[1]''. | ||
<br><br><br> | <br><br><br> | ||
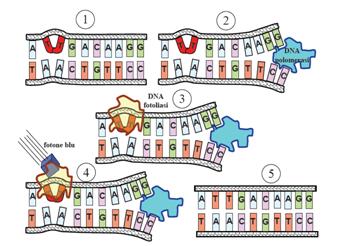
| - | This systems can be divided into two class: the SOS Photoreactivation repair and the SOS triggered by RecA protein. The first uses the photolyase, a poorly expressed enzyme(encoded by genes phrA and phrB) which binds the pyrimidine dimers and uses the blue light to split them apart as showed in Fig.2. | + | This systems can be divided into two class: the SOS Photoreactivation repair and the SOS respond triggered by RecA protein. The first uses the photolyase, a poorly expressed enzyme(encoded by genes phrA and phrB) which binds the pyrimidine dimers and uses the blue light to split them apart as showed in Fig.2. |
<br><br><br><br><br><br> | <br><br><br><br><br><br> | ||
| - | [[Image:fosfoliasi.jpg|300px|thumbnail|Fig.2|left]] | + | [[Image:fosfoliasi.jpg|300px|thumbnail|Fig.2 After dimerization of two adjacent thymines blue light gives energy to photolyase to repair the damage |left]] |
Revision as of 08:52, 29 October 2008
| HOME | PROJECT | TEAM | SOFTWARE | MODELING | WET LAB | SUBMITTED PARTS | BIOSAFETY AND PROTOCOLS |
|---|
Contents |
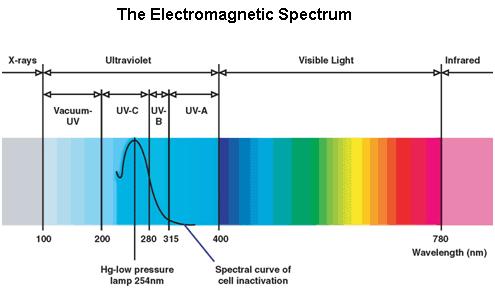
UV Spectrum
Ultraviolet is that part of electromagnetic radiation bounded by the lower wavelength extreme of the visible spectrum and the upper end of the X-ray radiation band. The spectral range of ultraviolet radiation is, by definition, between 100 and 400 nm and is invisible to human eyes. The UV spectrum is subdivided into three bands: UV-A (long-wave) from 315 to 400 nm, UV-B (medium-wave) from 280 to 315 nm, UV-C (short-wave) from 100 to 280 nm. A strong germicidal effect is provided by the radiation in the short-wave UV-C band.
E.coli SOS System
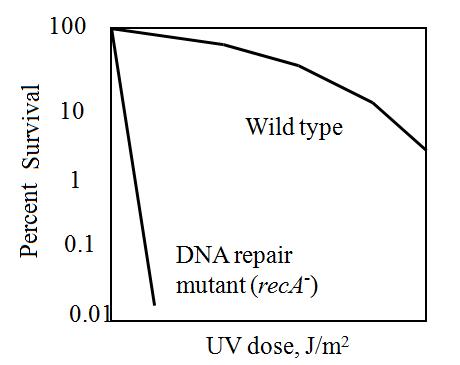
The maximum UV germicidal effect coincides with the peak absorbance of DNA (near 260nm) due to the dimerization of two adjacent thymines. That can be seen in the Fig.1 where is showed the living population of bacteria after irradiation.
E.Coli cells have a system that recovery DNA damage when it occurs and it is the best studied transcriptional response [1].
This systems can be divided into two class: the SOS Photoreactivation repair and the SOS respond triggered by RecA protein. The first uses the photolyase, a poorly expressed enzyme(encoded by genes phrA and phrB) which binds the pyrimidine dimers and uses the blue light to split them apart as showed in Fig.2.
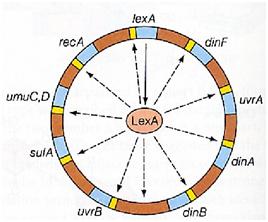
Instead single stranded DNA produced by several DNA-damaging agents can be bound by RecA protein, resulting in conversion of this protein to its activated form. The RecA repair system doesn’t need light and Lexa protein controls the expression of 43 genes [2] that cooperate together to repair the extensive genetic damage. RecA and LexA proteins play an important rule on the regulatory of SOS Recombination System.
A LexA binding site is present in all the SOS promoters genes' and it works as a repressor of SOS system. In presence of DNA damage (DNA Single Strains) RecA becomes active and interacts with LexA protein , the repressor of the SOS genes [3]. This interaction triggers the autocatalytic cleavage of LexA and consequent destruction of its ability to function as a repressor, which results in the derepression of SOS genes [4] [5]
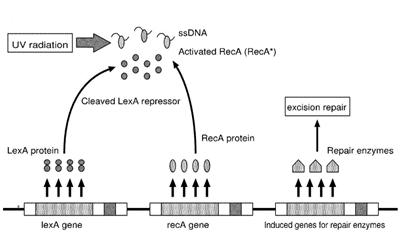
When the damage is repaired, DNA single strains are not present in the cell and the RecA protein no longer promotes the auto-cleavage of the LexA which is restored to its initial repression level.
LAB Experiment
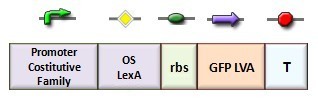
In our genetic bi-stable, the amount of molecules produced to switch the system from the two possible steady state is ruled by LacI and TetR protein. The production of LacI molecules by UV induction can be tested replacing the LacI gene by GFP, so it is possible to have a relation of the strength of LacI synthesis measuring the value of its fluorescence. BBa_K079049 and BBa_K079050 are two new construct submitted to the registry by Bologna’s Igem Team 2008. These UV test circuits can be presented schematically by the following sequence:
- Promoter Constitutive Family: BBa_J23118 (1429 strength ) or BBa_23100 (2547 strength);
- LexA Operator Site Binding (K079040 ) ;
- Rbs with Green fluorescence Protein LVA and Terminator (BBa_I763020)
In order to generate the induction by Uv, a box with UV lamp was realized.
UV Side Effect
In addiction to the germicidal effect UV radiation can also cause erythema (reddening of the skin)
and conjunctivitis (inflammation of the mucous membranes of the eye). Because of this,exist limit
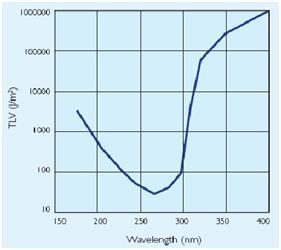
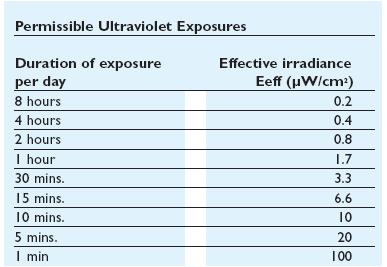
to the exposure[1]that depends by the irradiance of the ultraviolet- lamps(next figure).
Homemade UV transilluminator
When this device are used, it is important to design systems to exclude UV-C leakage and so avoid figure 4: Ultraviolet treshold limited value figure 5: Permissible ultraviolet exposure these effects.The UV source that have been used in this project is a GW6 Sylvania, a lamp that emitt UVC at 253,7nm near the absorption's peak of DNA with an optical output power of 1,6W; to use it safely we built a box of mdf(medium density-fibreboard) that surrounds the lamp and using a diaphragm that allows passage only to the light absorbing the remainder. Moreover we insert in that box two pierced brackets, those permits to choose the distance between the lamp and the sample and then in accord with the Lambert-Beer law's the exposure time.
Finally we embedded this structure in a polistyrene box for its handling and greater safety.(figure 6)
Gel matrix
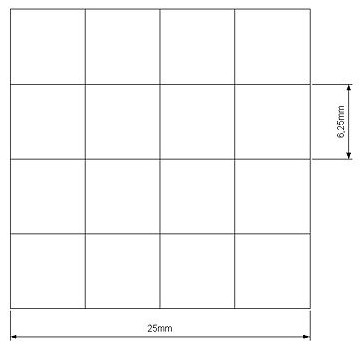
Our project use UV light for its space selectivity, that gives the possibility to irradiate a target zone without interfering with the other. To do that we build a mold to make a matrix of agarose gel;this give us a square of 25mm side's with 16 cells inside where we can locate bacteria.
Once do that is easy with an optical mask, like those used in photolithography for electronics
circuits, stimulate only the selected bacteria.To realize this device we use palsticard because it is
very easy to shape and clean.
The mold is made of two parts that will form a sandwich with the slide: one will create the shape
of the gel matrix the other is a cover that that will give the picture into gel .
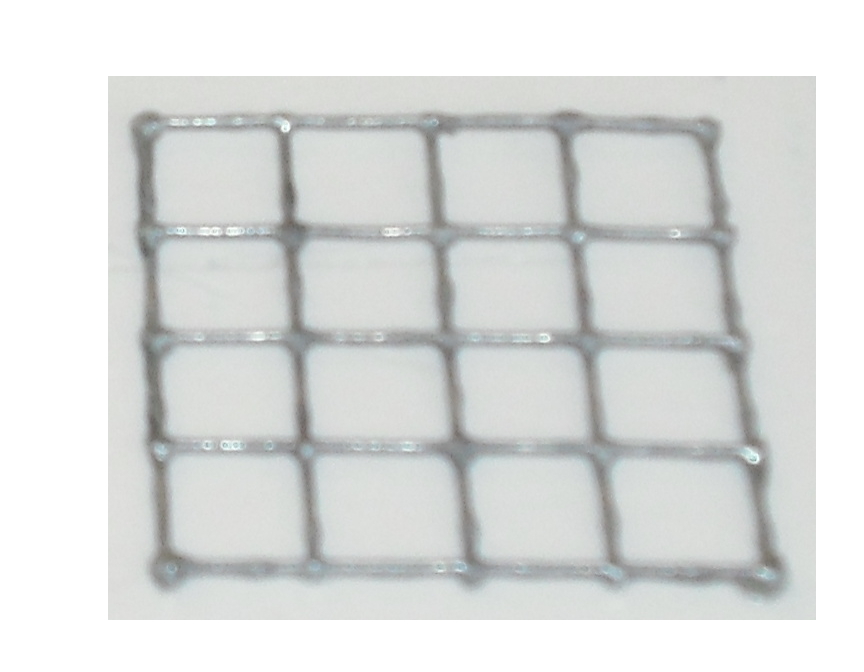
The matrix is obtained through the pressure of the cover part over the mold part
and that is the result:
Protocol used for UV induction
The plasmid with our construct has been transformed in DH5alfa bacteria by standard protocol and one colony from the plate has been picked and cultured overnight in 5ml LB medium broth with ampicilline.
The day after the culture has been diluted in 5ml LB and antibiotic in order to have 0.1 starting OD bacteria culture and let it grown for one hour; after that the culture has been divided in five falcons with 1ml of bacteria, ready to be induced. Using 1ml of culture is a choice done in order to have a thickness as thick as possible to perform an irradiation as uniform as possible .
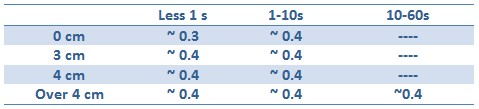
Tests with difference distances from the lamp with different exposition times were done in order to respect the maximum lethal UV dose of bacteria and avoid mutagenesis factors in the cell. The following table illustrates the tests setup used:
After induction by UV the samples were kept in dark by silver paper to increase the RecA and LexA answer and let grown for 2h. The OD sample is measured and the sample transferred in a 1ml eppendorf spinned at 6000-8000rpm for three minutes and pellet collected in order to measure fluorescence level by microscope and Bacteria Visual Fluo Software.
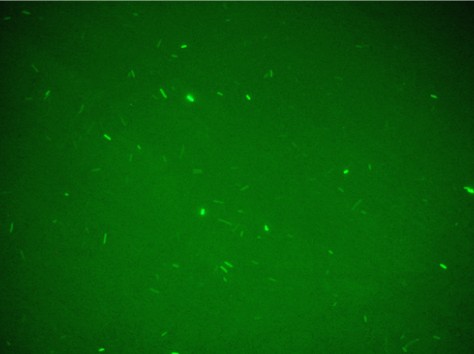
Unfortunately, using Falcon tube and not the correct time/distance induction the experimental tests showed weak GFP expression using the construct with straighter promoter. It could be depended by the low sensibility of SOS system with our setup tests and not uniform irradiation of the sample. In Fig.3 is possible to see a leak answer by bacteria stimulated by UV at the distance of 4cm with one second time exposure.
In order to show the correct functionality of our construction, induction by peroxide hydrogen was used, in fact a low concentration of H_2 O_2 (1-3mM) results in SOS gene induction in wild-type cells [6], [7].
The induction of SOS by H_2 O_2 is an important event, since RecA and recBC mutant cells are very sensitive to H_2 O_2 treatment probably due to the lack of recombinational repair necessary for the H_2 O_2 – induced lesions [8].
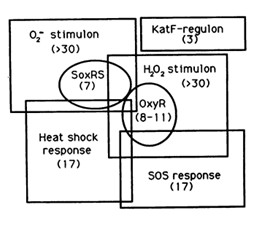
The reaction of hydrogen peroxide with transition metals imposes on cells an oxidative stress condition that can result in damage to cell components such as proteins, lipids and principally to DNA. Escherichia Coli cells are able to deal with these adverse events via DNA repair mechanisms or OxyR and SosRS anti-oxidant inducible pathways which are elicited by cells to avoid the introduction of oxidative lesions by hydrogen peroxide.
Among the systems the OxyR gene interacts with, there is the SOS response.
Protocol Used for Hydrogen Peroxide induction
The plasmid contained the LexA Operator was transformed into DH5alfa bacteria by standard protocol and one colony has been picked from plate and let it grown overnight in Falcon with 5ml LB broth with ampicilline.
Cultured colony has been diluted in 5ml LB broth with ampicilline and 1ul H2O2 (11M) to have medium with 2.2mM H202 and 0.1 starting OD. A test sample culture has been obtained diluting at the same way in LB medium and antibiotic without H202.
Both falcon were kept to the dark by using silver paper and let them growing for 2 hours at 37°C. 1 ml of bacteria sample has been collected and transferred inside 1ml eppendorf and spinned at 6000-8000 rpm for 3 minutes. The supernatant has been throw away and the pellet resuspended for the slide preparation. This procedure gave the uniformity of induction to all the sample and the Fig.4 shows that GFP synthesis is started in all the bacteria population.
In order to prove that these constructs can be used in the bi-stable circuit without interference between Lac Operon and SOS System, a test with LexA site binding construct and 256Promoter + Laci + T + Plac + RBS + GFP LVA was used. The first and the latter construct were induced by H2O2 or IPTG and in Fig.5 is possible to see the results.
As expected the Lexa construct induced by IPTG and Lac-Operon-based-circuit induced by H2O2 didn’t produced GFP synthesis, instead the other two sample correctly induced presented good level of fluorescence.
[1] Friedberg EC, Walker GC and Siede W (1995) DNA Repair and Mutagenesis. Academic Press, New York, 698 pp
[2] Courcelle J, Khoudursky A, Peter B, Brown PO and Hanawalt PC (2001) Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64
[3] Wagner J, Cruz P, Kim SR, Yamada M, Matsui K, Fuchs RP and Nohmi T (1999) The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell 4:281-286
[4] Mustard JA and Little JW (2000) Analysis of Escherichia coli RecA interactions with LexA, lambda CI, and UmuD by site-directed mutagenesis of rec
[5] Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H and Woodgate R (2000)
Identification of additional genes belonging to the lexA regulon in Escherichia coli. Mol Microbiol 35:1560-1572
[6] Imlay JA and Linn S (1987) Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bateriol 169:2967-2976
[7] Goerlich O, Quillardet P and Hofnung M (1989) Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J Bacteriol 171:6141-6147
[8] Imlay JA and Linn S (1987) Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide
 "
"