Team:Newcastle University/Software
From 2008.igem.org
| Line 5: | Line 5: | ||
==Components== | ==Components== | ||
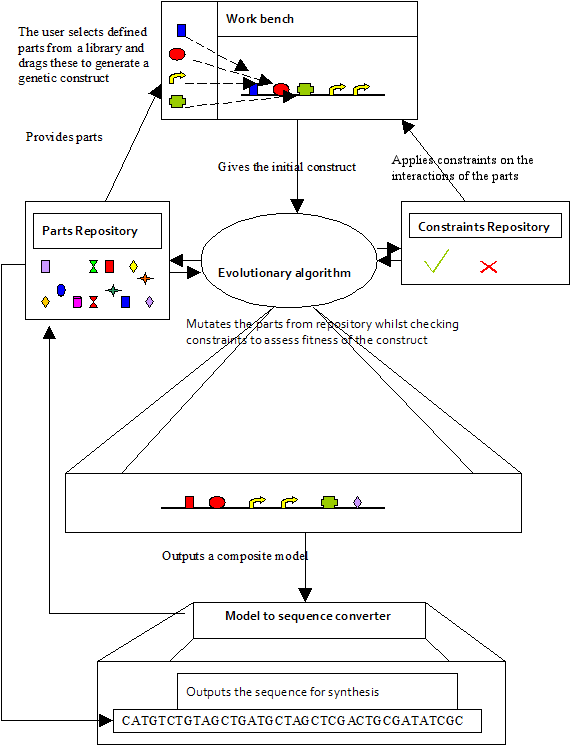
| - | [[Image:Ncl-software.png|thumb|left|500px| | + | [[Image:Ncl-software.png|thumb|left|500px|Schematic of the components of the software system. The workbench contains parts provided by the parts repository, and constraints provided by the constraints repository. Parts are selected by the user to create an initial structure for the genetic circuit. Constraints define whether these parts can fit together. The workbench outputs this user defined structure to the evolutionary algorithm, which evolves the circuit by requesting different parts from the parts repository, and generates a fitness value of the mutated circuit using values provided by the constraints repository. Using part model s provided a composite model could be formed. The fittest composite model is output to the model to sequence converter, which uses the parts to derive the sequence of the construct and how they fit together to give an overall nucleotide sequence.]] |
===Workbench (Morgan)=== | ===Workbench (Morgan)=== | ||
Revision as of 17:29, 28 October 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Software
Our software consists of four main modules which communicate via Web services. One person was Lead Developer for each module, but the developers worked in the same lab, and had frequent formal and informal discussions in order to develop software which works together and provides the necessary functionality.
Components
Workbench (Morgan)
The Workbench is a graphical front end that allows circuits to be designed by hand, using drag-and-drop of icons, and drawing upon the Parts Repository and the Constraints Repository. The designed circuits can then be sent to the EA to be further refined.
Parts Repository (Megan)
The Parts Repository stores information from the literature and various databases about biological parts such as promoters, ribosomal binding sites and protein coding regions.
Constraints Repository (Nina)
The Constraints Repository stores information about how parts work together, and associated parameters such as binding affinities, POPS, etc., where these are known. Populated from the literature and hard-won knowledge from advisors.
Evolutionary Algorithm (Mark)
The EA takes parts from the Parts Repository, assembles them randomly, but using constraints from the Constraints Repository, and evaluates the "fitness" of the resulting circuit depending on a desired input:output mapping. Equivalent parts with different parameters are swapped in and out at random, and fitter circuits selected in an iterative manner.
 "
"

