Newcastle University Wetlab/5 September 2008
From 2008.igem.org
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Wet Lab >> Newcastle Wet Lab Journal
Wet lab work was carried out from 4 August to 19 September, Mondays to Fridays. Please click on a day to see the lab notebook.
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Friday 05th September
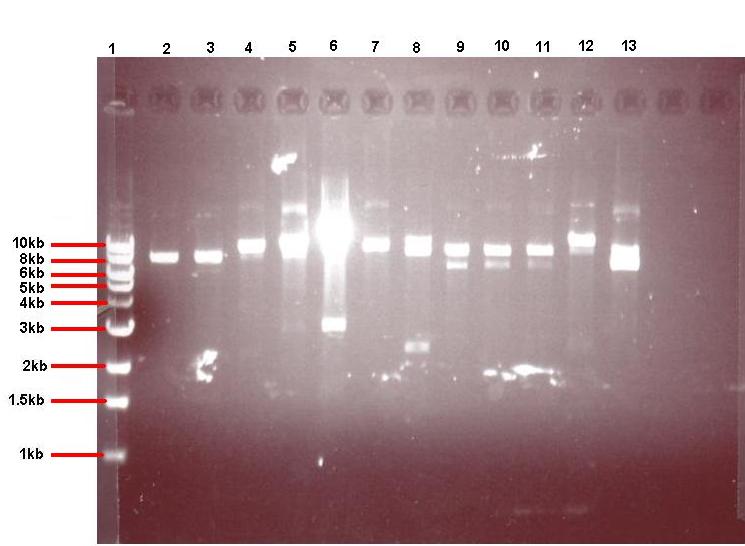
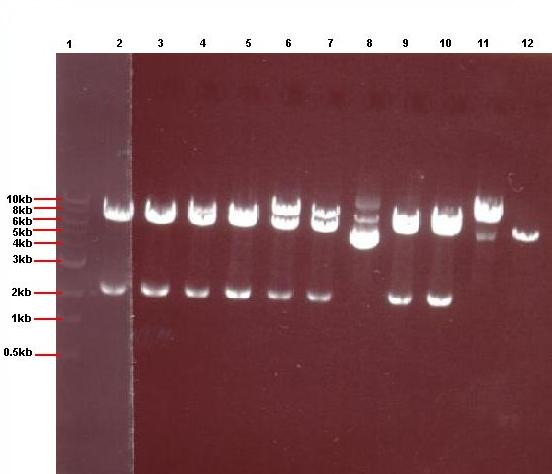
- 4μl of each of the 24 overnight cultures was centrifuged and the plasmid isolated from the resulting pellets. The isolated plasmid was then restricted to see if the plasmid had taken up the insert. All pGFPrrnB-nvl08 plasmids were restricted using EcoRI and NheI and pJWV021-ncl08 plasmids restricted with NheI and BglII. The restrictions were carried out in using 7.5μl plasmid in a total volume of 15μl. Samples were run on gel.
Lane 1: 1kb ladder
Lane 2: pGFPrrnB-ncl08 colony 1 (white colony, 25μl plate)
Lane 3: pGFPrrnB-ncl08 colony 2 (white colony, 25μl plate)
Lane 4: pGFPrrnB-ncl08 colony 3 (white colony, 25μl plate)
Lane 5: pGFPrrnB-ncl08 colony 4 (white colony, 25μl plate)
Lane 6: pGFPrrnB-ncl08 colony 5 (white colony, 25μl plate)
Lane 7: pGFPrrnB-ncl08 colony 6 (white colony, 25μl plate)
Lane 8: pGFPrrnB-ncl08 colony 7 (white colony, 25μl plate)
Lane 9: pGFPrrnB-ncl08 colony 8 (white colony, 25μl plate)
Lane 10: pGFPrrnB-ncl08 colony 9 (white colony, 25μl plate)
Lane 11: pGFPrrnB-ncl08 colony 10 (white colony, 250μl plate)
Lane 12: pGFPrrnB-ncl08 colony 11 (white colony, 250μl plate)
Lane 13: pGFPrrnB-ncl08 colony 12 (green colony, 25μl plate)
Lane 1: 1kb ladder
Lane 2: pJWV021-ncl08 colony 1 (white colony)
Lane 3: pJWV021-ncl08 colony 2 (white colony)
Lane 4: pJWV021-ncl08 colony 3 (white colony)
Lane 5: pJWV021-ncl08 colony 4 (white colony)
Lane 6: pJWV021-ncl08 colony 5 (white colony)
Lane 7: pJWV021-ncl08 colony 6 (white colony)
Lane 8: pJWV021-ncl08 colony 7 (white colony)
Lane 9: pJWV021-ncl08 colony 8 (white colony)
Lane 10: pJWV021-ncl08 colony 9 (white colony)
Lane 11: pJWV021-ncl08 colony 10 (white colony)
Lane 12: pJWV021-ncl08 colony 11 (white colony)
Lane 13: pJWV021-ncl08 colony 12 (white colony)
- The gels showed correct insert fragments (2.2kb) in lanes 8 (colony 7) and 12 (colony 11) for pGFPrrnB and lanes 2, 3, 4, 5, 6, 7, 9 and 10 (colonies 13, 14, 15, 16, 17, 18, 20 and 21 respectively) for pJWV021.
- Unrestricted plasmid from colonies 7 + 11 (pGFPrrnB) and 13 + 14 (pJWV021) were transformed into Bacillus subtilis (see Transforming into Bacillus subtilis).
- Plasmid renamed once transformed into B. subtilis to avoid confusion with plasmid in E. coli:
- pGFPrrnB-ncl08 (E. coli) = iGEMgfp (B. subtilis)
- pJWV021-ncl08 (E. coli) = iGEMcherry (B. subtilis)
- Agar cultures were also made from the same colony cultures:
- iGEMgfp (chloramphenicol) colony 7
- iGEMgfp (chloramphenicol) colony 11
- iGEMgfp (chloramphenicol) negative control
- iGEMcherry (kanomycin) colony 13
- iGEMcherry (kanomycin) colony 14
- iGEMcherry (kanomycin) negative control
 "
"