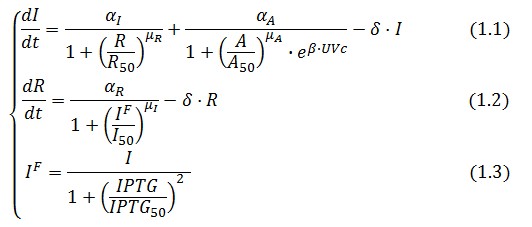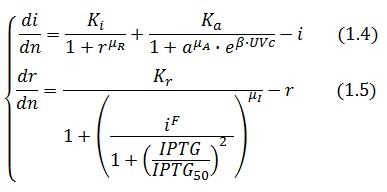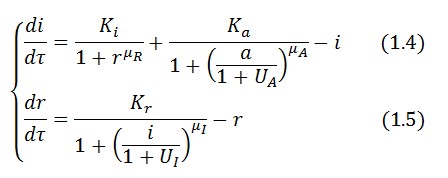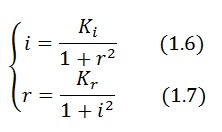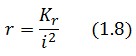Team:Bologna/Modeling
From 2008.igem.org
| HOME | TEAM | PROJECT | MODELING | WET-LAB | SOFTWARE | SUBMITTED PARTS | BIOSAFETY AND PROTOCOLS |
|---|
Contents |
Model-based analysis of the genetic Flip-Flop
The genetic Flip-Flop
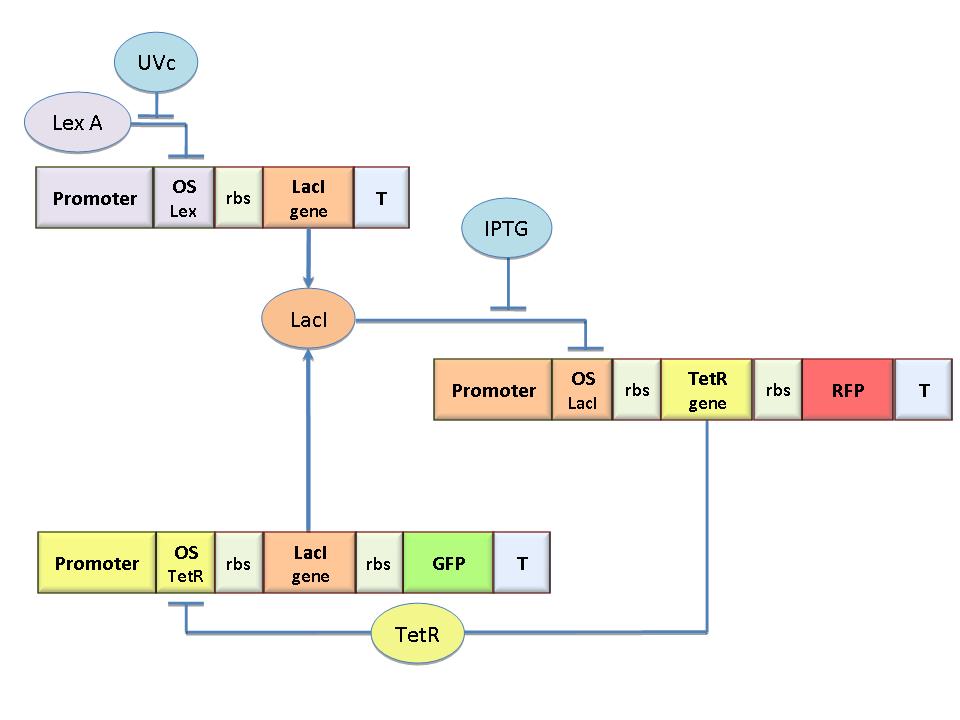
The molecular circuit in Figure 1 works as a Flip-Flop (SR Latch), which can switch between two different states according to the external stimuli (UVc and IPTG).
LacI-ON represents the stable state in which LacI gene is active and LacI protein represses the TetR gene expression, with a positive feedback.
Therefore, the LacI-ON state coincides with the TetR-OFF condition. On the contrary, the TetR-ON represents the state with the TetR gene active and the LacI gene silenced (LacI-OFF). Owing to the coexistence of two stable states (bistability), this circuit is capable of serving as a binary of memory. We denominated it Flip-Flop since it works as a SR Latch: LacI state is the output and TetR state is the output. Uvc is the set signal and IPTG is the reset signal. Indeed, IPTG stimulation inhibits LacI repressor, thus can cause the transition from the LacI-ON state to the TetR-ON, whereas UVc radiation inactiving LexA repressor through the SOS response (Friedberg et al., 1995) can cause the opposite transition from LacI-ON to TetR-ON.
Mathematical model
Model equations
The Flip-Flop circuit in Figure 1 can be modeled by the following equations:
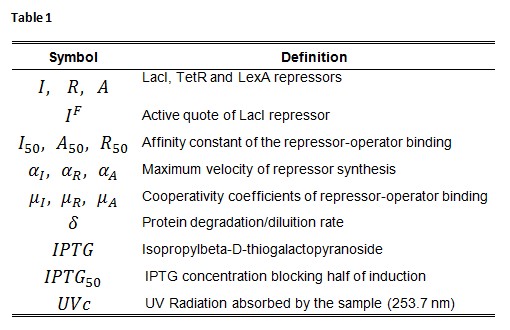
Symbol definition is listed in Table 1.
A common motif in repressor proteins is the presence of a dimeric nucleotide-binding site with dimeric structure. In accordance to this general structure the cooperativity coefficients ( ) were assumed equal to 2. The maximum velocity of repressor synthesis ( accounts for the strength of the unregulated promoter and RBS. The value of the affinity constant for the binding of repressor to the promoter strictly depends on the sequence of operator site (OS block).
Adimensional equations
The equations (1.1) and (1.2) can be written dimensionless:
Adimensional equations
The equations (1.1) and (1.2) were modified to an adimensional form:
Equibrium conditions
In the absence of stimuli, the adimensional concentrations of LacI ( ) and TetR ( ) at equilibrium are related by the equations:
To obtain these relations the UVc-dependent term in equation (1.4) was ignored ( ). This is justified by the high binding constant of LexA for its operator, and the consequent negligible contribution to the LacI synthesis. Equations (1.6) and (1.7) can have one or three solutions that represent the equilibrium conditions of the circuit. The solutions, i.e. the equilibrium conditions, can be graphically identified as the intersections between r and i nullclines (see Figure 2). The case of multiple equilibrium conditions (bistability case) is shown in Figure 2 panel b). TetR-ON and LacI-ON are stable equilibriums separated by the unstable one (saddle point). Due to the bistability the circuit can operate as a binary memory. The existence of a bistability condition depends on the value of Kr and Ki parameters. If Kr decrease (see Figure 2 pannel (a)) a saddle-node bifurcation can occur, TetR-ON equilibrium vanishes and remain only the stable equilibrium LacI-ON. The contrary occurs when Ki is decreased (Figure 2 pannel (c)). Thus bistability is guarantied only for a limited range of and values.
Bifurcation analysis
Assuming that LacI-ON exists the corresponding equilibrium value of is higher than 1 (see Figure 2), then it can be assumed that and the equation (1.7) simplies to:
Substituting this expression in equation (1.6) one obtain:
[[Image:equa1_9.jpg|center]
Stability analysis
Assuming as starting condition the LacION state (Figure 1) its stability is guaranteed if the adimensional concentration of LacI is higher than 1 (Figure 2).
In the limit of (), equation (1.7) simplifies:
Experimental identification
The MAVs were identified comparing the experimental response of the genetic circuits in Figure
 "
"