Team:Bologna/Project
From 2008.igem.org
| HOME | TEAM | PROJECT | MODELING | WET-LAB | SOFTWARE | SUBMITTED PARTS | BIOSAFETY AND PROTOCOLS |
|---|
Contents |
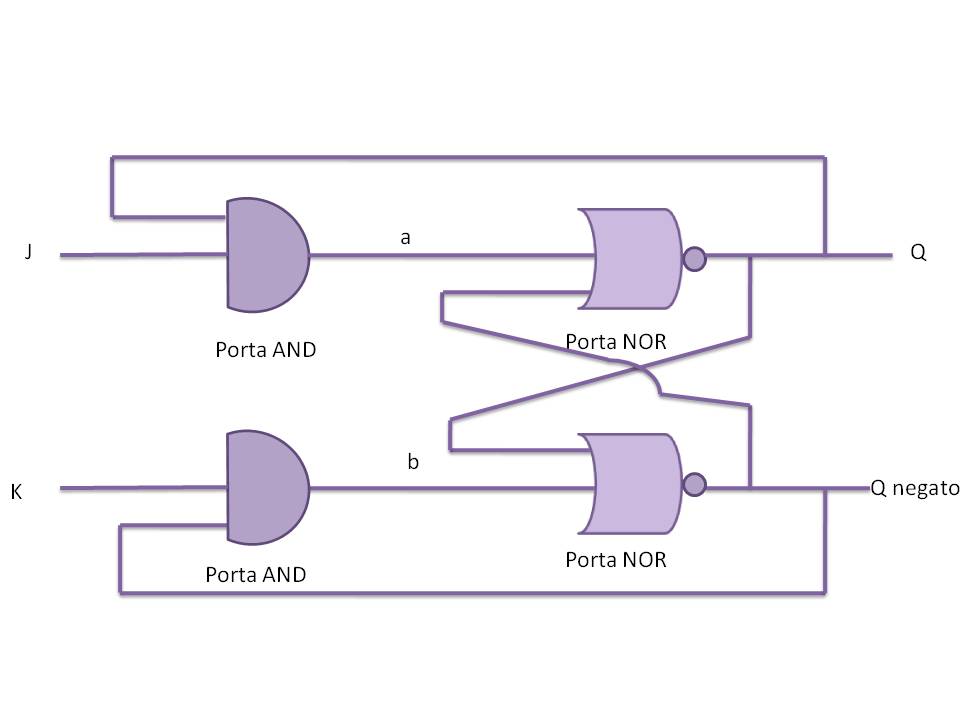
Flip flop logic circuit
Ecoli.PROM: an Eresable and Programmable Genetic Memory in E. coli
The specific goal of our project was to design a bacterial reprogrammable memory, i.e. colonies of genetically engineered E. coli immobilized in solid medium where they work as an array of binary memory cells.
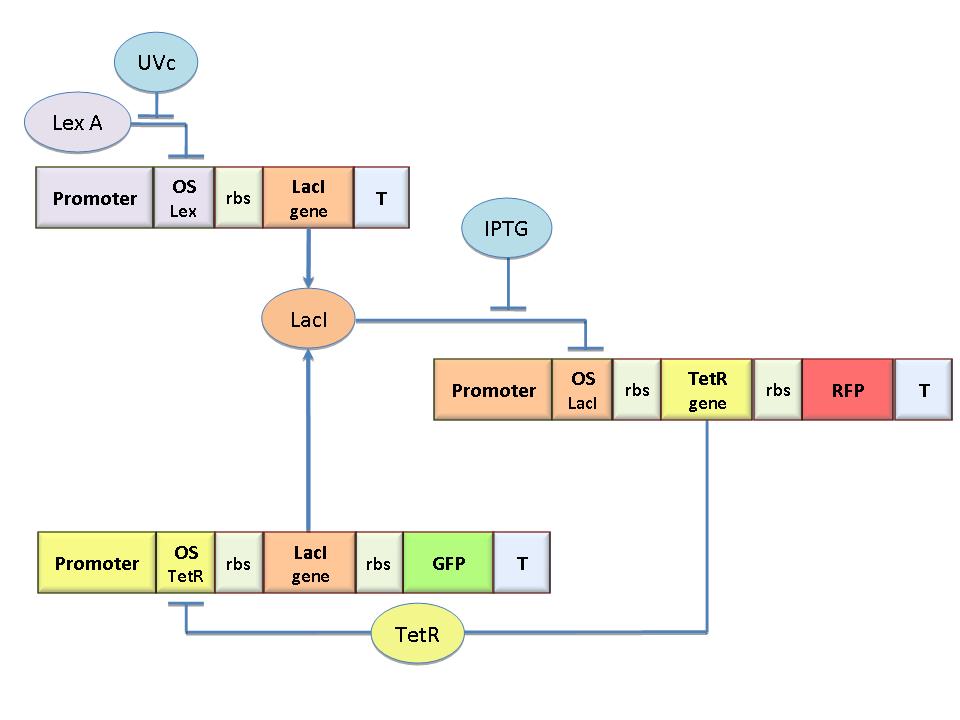
To engineer the bacteria we designed a modular genetic flip-flop composed of two parts: a binary memory block (toggle switch) and an induction block sensitive to UV radiation to set ON the memory. UV has been chosen to have a fine spatial selectivity in programming the memory cells, whereas IPTG should be used to reset the entire memory. Since UV radiation is detrimental for the cells, we have designed a circuit with very high UV sensivity to limit the exposition time. The fine tuning of the circuit elements was achieved by a model-based analisys.
The core elements of the genetic memory are two mutually regulated promoters each designed as indipendent operator sites flanking a constitutive promoter. In this way the promoter transcriptional strength and the repressor binding affinity can be independently fixed. To this aim we designed operator libraries for LacI, TetR, Lambda and LexA repressor cloning them in the BioBrick format for their standard assembly. This will allow the rational design of regulated promoter elements that are still lacking in the Registry.
This approach has been applied in the building of our UV-programmable memory, and we expect it to be a general benefit in a larger number of applications in Synthetic Biology.
Collaboration with other iGEM Teams 2008
- After the last year competition, at the beginning of the 2008, we decided to get a new team started for iGEM2008 competition. In April, we got in contact with Prof. Paolo Magni, who wanted to start a new team in Pavia for iGEM2008. So, in order to share experiences and ideas about iGEM, and to show him what kind of wet lab resources are necessary to develop a Synthetic Biology project, we met at the Cellular and Molecular Engineering Laboratory of the University of Bologna- Cesena Campus. After this first meeting, there have been other chances to meet during the summer. In particular, several conference calls were organized and two meetings were scheduled in Pisa and Bressanone (Italy). It was fundamental to compare lab protocols and techniques to help each other avoiding mistakes and speeding up project progress. The main topics of our discussion were the optimization of plasmid resuspension and ligation reaction steps as well as how to measure fluorescence. Finally, before DNA Repository quality control publication on the Registry web site, we cross-checked some parts that showed problems after DNA transformation. Problems had been confirmed by quality control results (parts' sequences classified as "inconsistent").
- We want even to mention the courtesy of the Valencia iGEM Team, that have advised us about the critical use of GFP and RFP at the same time.
Project Details
In this paragraph you can find some information about main project topics.
 "
"