Team:Calgary Wetware/Project
From 2008.igem.org
(Prototype team page) |
(→Applications) |
||
| (40 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
!align="center"|[[Team:Calgary_Wetware|Home]] | !align="center"|[[Team:Calgary_Wetware|Home]] | ||
!align="center"|[[Team:Calgary_Wetware/Team|The Team]] | !align="center"|[[Team:Calgary_Wetware/Team|The Team]] | ||
!align="center"|[[Team:Calgary_Wetware/Project|The Project]] | !align="center"|[[Team:Calgary_Wetware/Project|The Project]] | ||
| - | |||
| - | |||
!align="center"|[[Team:Calgary_Wetware/Notebook|Notebook]] | !align="center"|[[Team:Calgary_Wetware/Notebook|Notebook]] | ||
| + | !align="center"|[[Team:Calgary_Wetware/Protocols|Protocols]] | ||
| + | !align="center"|[[Team:Calgary_Wetware/Results|Results]] | ||
| + | !align="center"|[[Team:Calgary_Wetware/Beyond|Beyond the Lab]] | ||
| + | !align="center"|[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Calgary_Wetware Parts] | ||
|} | |} | ||
| - | + | <br><br><br> | |
| + | [[image:Batman4.gif|93px]] [[image:BadGuy1.gif|105px]][[image:BadGuy2.gif|105px]] | ||
| + | == '''Overall Project''' == | ||
| - | + | Microorganisms use pheromones to monitor their own population density as well as to detect and interact with other microbial species in a process known as quorum sensing. For instance, bacteria can disrupt the pheromone signals of other competing bacteria, effectively preventing their proliferation. In a similar sense, we will exploit the natural communication systems involving Autoinducer-1 (AI-1) from ''Vibrio fischeri'' and Autoinducer-2 (AI-2) from ''Vibrio harveyi'', to create a model biosensor system in ''Escherichia coli''. We will engineer the genetic circuits necessary for the production of these pheromones into two populations of ''E. coli'' (termed Bad guy #1 and Bad guy #2, as per their respective Autoinducer). In addition, our third population of ''E. coli'' (termed Champion cell) acts as a biosensor by receiving these signal inputs and subsequently initiating transcription of specific ''E. coli''-targeted bacteriocins (''i.e.'' colicins). The presence of AI-1 induces the Champion to produce a colicin to which Bad guy #1 is susceptible, but to which Bad guy #2 is resistant, and vice-versa for AI-2. An additional aspect of our system is the ability of the Champion cell to report the presence of each specific Bad guy by producing a specific fluorescent protein (''i.e.'' either green or red fluorescent protein) in tandem with the specific colicin as determined by the presence of either AI-1 or AI-2. We constructed this system using the molecular cloning methods utilized in iGEM. While the ''Vibrio fischeri'' components of our system were obtained as standardized parts from the iGEM Registry, we plan to clone and standardize all parts of ''Vibrio harveyi'' AI-2 quorum sensing system. All of the standardized parts will be flanked by the specific restriction endonuclease sites as required for iGEM BioBricks. This will allow for the simple and iterative directional cloning strategy for the construction of the necessary genetic circuits. | |
| - | + | == '''System #1 In Detail''' == | |
| + | [[image:BadGuy1.gif|35px]] | ||
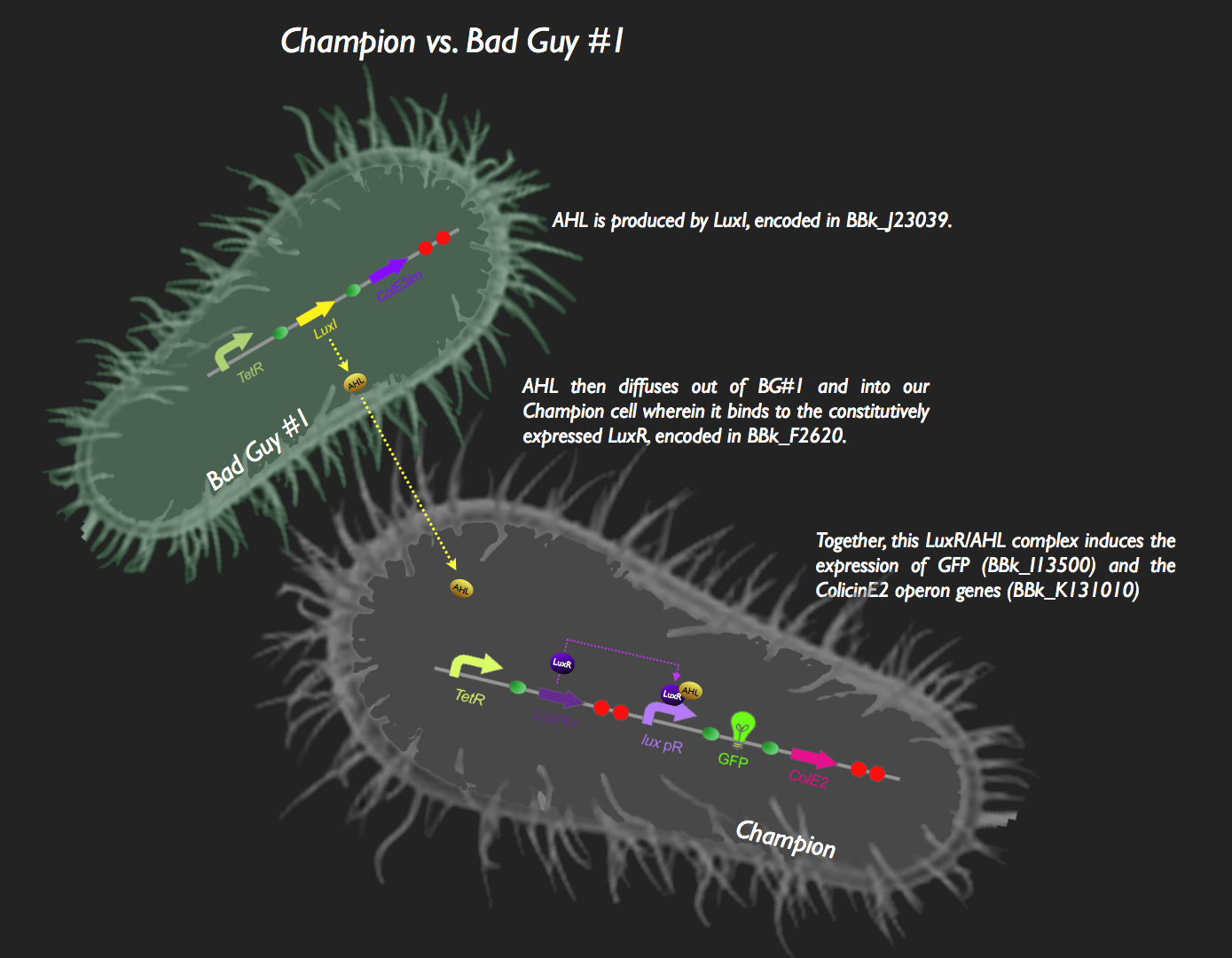
| + | The first half of our system has Bad Guy #1 as the culprit. Bad Guy #1 is capable of producing large amounts of autoinducer-1, more commonly known as Acyl Homoserine Lactone. This is through constitutive expression of the ''luxI'' gene. AHL begins to diffuse into our Champion Cell, which is constitutively expressing LuxR. LuxR protein binds to the pheromone, and the AHL-LuxR complex activates the lux pR promoter, allowing the expression of green fluorescent protein and the colicin E2 activity, immunity and lysis genes (CeaB, CeiB and CelB respectively). The champion cell now fluoresces green, and colicin expression results in the lysis of the cell and the release of CeaB, which would be taken up by Bad Guy #1. Colicin E2 would lead to the eventual destruction of Bad Guy #1, and our Champion Cell will reign supreme (those that haven't burst from the production of the Colicin proteins and are still kicking around, anyway). | ||
| + | [[image:Sys1.jpg|800px]] | ||
| + | This system was assembled in typical iGEM fashion. The part [http://partsregistry.org/Part:BBa_J23039 J23039] was transformed into ''E. coli'' thus resulting in Bad Guy #1 cells (i.e. capable of AHL production). To construct the signaling and response circuit in the Champion Cell, [http://partsregistry.org/Part:BBa_F2620 F2620], [http://partsregistry.org/Part:BBa_I13500 I13500] and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K131009 colicin E2 operon] were assembled as shown in the picture. Colicin E2 was obtained from UC Berkeley, cloned into a TOPO vector (Invitrogen), amplified using ColicinE2 primers with flanking Biobrick Restriction Sites and cloned into a Biobrick vector. | ||
| + | Tests were also performed, the results of which can be seen in the [https://2008.igem.org/Team:Calgary_Wetware/Results Results] section. These involved verifying that Bad Guy #1 is able to make AHL, verifying that our Champion Cell responds to AHL produced from Bad Guy #1, and that Colicin E2 was actually capable of preventing bacterial growth. | ||
| + | <br><br> | ||
| + | <html> | ||
| + | <div style="margin:10px"> | ||
| + | <object width="765" height="600"> | ||
| + | <param name="movie" value="https://static.igem.org/mediawiki/2008/4/4a/System1_movie.swf"> | ||
| + | </param> | ||
| + | <param name="wmode" value="transparent"> | ||
| + | </param> | ||
| + | <embed src="https://static.igem.org/mediawiki/2008/4/4a/System1_movie.swf" type="application/x-shockwave-flash" wmode="transparent" width="765" height="600" ></embed> | ||
| + | </object> | ||
| + | </div> | ||
| + | </html> | ||
| + | ''Click on the animation above to watch our system in action.'' | ||
| + | <br><br> | ||
| - | == | + | == '''System #2 In Detail''' == |
| + | [[image:BadGuy2.gif|35px]] The other half of our system sees the aptly named Bad Guy #2 as the main adversary to our Champion Cell. | ||
| + | [[image:Sys2.jpg|800px]] | ||
| + | <br><br> | ||
| + | Without any AI-2 in the vicinity, however, there is downstream expression of a repressor protein, clλ, by phosphorylated LuxO, thereby masking the expression of ColicinE3 and RFP. Otherwise, without the presence of this inverter, our Champion cell would start mass production of ColE3 and RFP (indicating the presence of the villain) in the absence of any threat... the '''exact opposite''' of what we want!! | ||
| + | <html> | ||
| + | <div style="margin:10px"> | ||
| + | <object width="765" height="600"> | ||
| + | <param name="movie" value="https://static.igem.org/mediawiki/2008/3/35/System_2_NO_AI2.swf"> | ||
| + | </param> | ||
| + | <param name="wmode" value="transparent"> | ||
| + | </param> | ||
| + | <embed src="https://static.igem.org/mediawiki/2008/3/35/System_2_NO_AI2.swf" type="application/x-shockwave-flash" wmode="transparent" width="765" height="600" ></embed> | ||
| + | </object> | ||
| + | </div> | ||
| + | </html> | ||
| + | ''Click on the animation above to see what happens when the cell doesn't make anything.'' | ||
| + | <br><br> | ||
| + | But then bad Guy #2 comes along. He’s generating AI-2 all by himself, without any engineering necessary (as a result of constitutive expression of ''luxS'', encoding the LuxS enzyme which produces an intermediate in the AI-2 biosynthesis pathway) which is intercepted by the periplasmic protein LuxP in the Champion Cell which is constitutively bound to the LuxQ protein, a histidine kinase spanning the periplasmic membrane. The binding of AI-2 to LuxP induces the stabilization of the phosphatase conformation of the cytosolic domain of LuxQ, thereby allowing for the dephosphorylation of LuxU. The loss of phosphorylation of LuxU puts an end to its kinase activity, with the net effect being the dephosphorylation of LuxO. Since phospho-LuxO is required for the activation of the ''qrr4'' promoter, no more clλ repressor is produced, resulting in the activation of the ''pclλ'' promoter thanks rapid destruction of the repressor afforded by the degradation tag attached to the repressor protein. ColicinE3 is transcribed, leading to the eventual destruction of Bad Guy #2, following the release of the cytotoxic protein from our champion cell. Concurrently, our Champion Cell fluoresces red to inform the viewing gallery who the invader is and in order to indicate that our champion cells are preparing to wage war!<br> | ||
| + | <html> | ||
| + | <div style="margin:10px"> | ||
| + | <object width="765" height="600"> | ||
| + | <param name="movie" value="https://static.igem.org/mediawiki/2008/8/8d/System_2_with_AI2.swf"> | ||
| + | </param> | ||
| + | <param name="wmode" value="transparent"> | ||
| + | </param> | ||
| + | <embed src="https://static.igem.org/mediawiki/2008/8/8d/System_2_with_AI2.swf" type="application/x-shockwave-flash" wmode="transparent" width="765" height="600" ></embed> | ||
| + | </object> | ||
| + | </div> | ||
| + | </html> | ||
| + | ''Click on the animation above to see what happens when our champion finally sees Bad Guy 2 and fights back!'' | ||
| + | <br><br> | ||
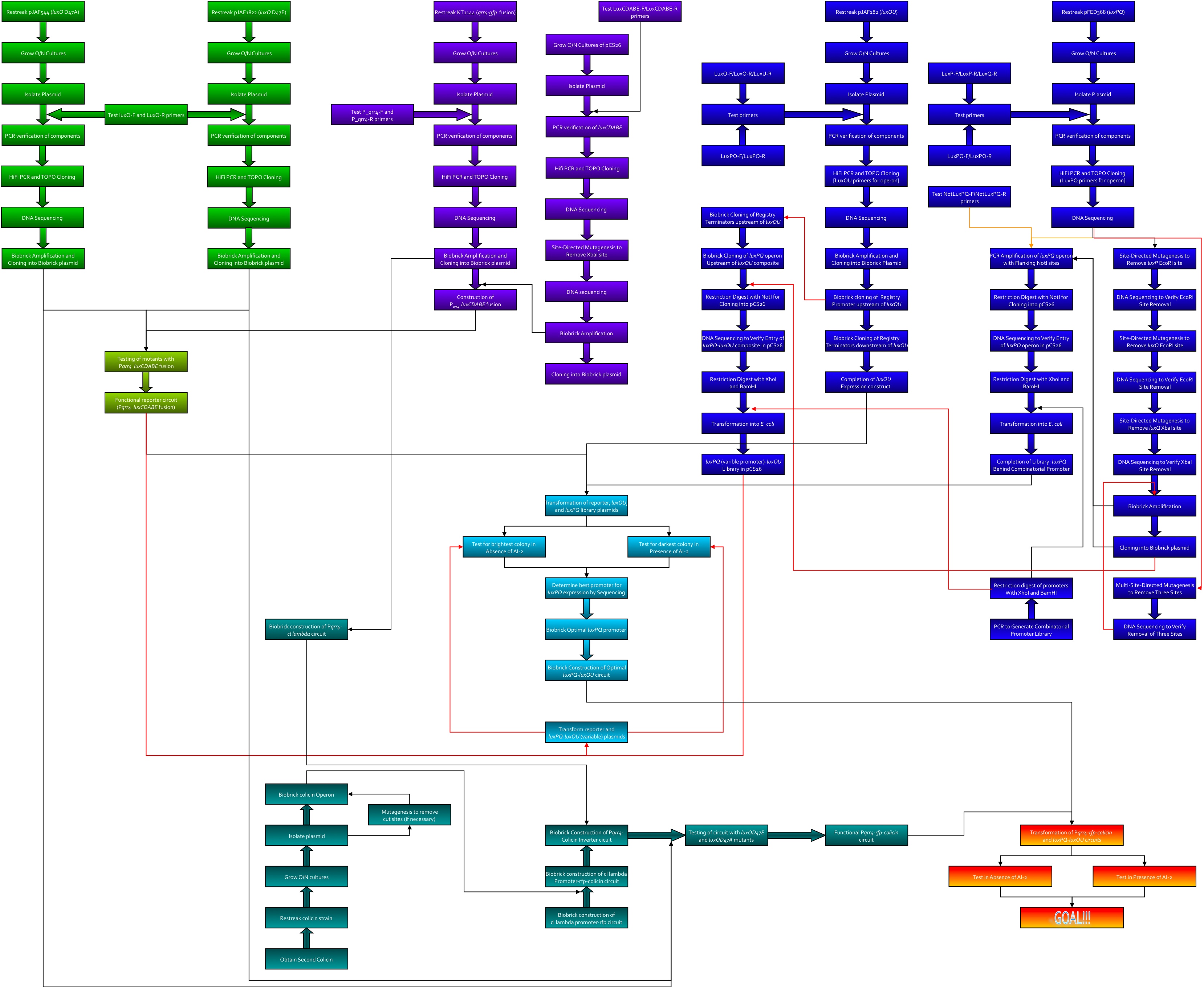
| - | + | Here's the workflow envisioned for creating system 2. Kingsley worked on the green section, Kevin was to work on the purple, Thane worked on the blue, and Dave worked on the teal.<br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[image:System2.jpg|965px]] | ||
| + | == '''Applications''' == | ||
| + | In essence, what’s proposed above is a model or proof of concept system. Then, where could this system be useful? One evident application stems from the knowledge that specific bacterial strains create bacteriocins targeted uniquely to their populations (''i.e. E. coli'' employ colicins while ''P. aeruginosa'' use pyocins). Combining the production of species-specific bacteriocins and reporter activity with the use of species-specific pheromone signaling systems would allow our Champion to recognize some of the real bad guys out there like ''Yersinia pestis'' (i.e., the pathogen responsible for causing Bubonic plague). | ||
| - | + | In addition, the contribution of a working of a second system for bacterial cell to cell communication (i.e., in addition to AI-1 signaling) within the Registry would drastically increase the range of future projects that could be undertaken by providing the users with an additional input (i.e. AI-2) for their genetically engineered machines. | |
Latest revision as of 03:10, 30 October 2008
| Home | The Team | The Project | Notebook | Protocols | Results | Beyond the Lab | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Calgary_Wetware Parts] |
|---|
Contents |
Overall Project
Microorganisms use pheromones to monitor their own population density as well as to detect and interact with other microbial species in a process known as quorum sensing. For instance, bacteria can disrupt the pheromone signals of other competing bacteria, effectively preventing their proliferation. In a similar sense, we will exploit the natural communication systems involving Autoinducer-1 (AI-1) from Vibrio fischeri and Autoinducer-2 (AI-2) from Vibrio harveyi, to create a model biosensor system in Escherichia coli. We will engineer the genetic circuits necessary for the production of these pheromones into two populations of E. coli (termed Bad guy #1 and Bad guy #2, as per their respective Autoinducer). In addition, our third population of E. coli (termed Champion cell) acts as a biosensor by receiving these signal inputs and subsequently initiating transcription of specific E. coli-targeted bacteriocins (i.e. colicins). The presence of AI-1 induces the Champion to produce a colicin to which Bad guy #1 is susceptible, but to which Bad guy #2 is resistant, and vice-versa for AI-2. An additional aspect of our system is the ability of the Champion cell to report the presence of each specific Bad guy by producing a specific fluorescent protein (i.e. either green or red fluorescent protein) in tandem with the specific colicin as determined by the presence of either AI-1 or AI-2. We constructed this system using the molecular cloning methods utilized in iGEM. While the Vibrio fischeri components of our system were obtained as standardized parts from the iGEM Registry, we plan to clone and standardize all parts of Vibrio harveyi AI-2 quorum sensing system. All of the standardized parts will be flanked by the specific restriction endonuclease sites as required for iGEM BioBricks. This will allow for the simple and iterative directional cloning strategy for the construction of the necessary genetic circuits.
System #1 In Detail
![]() The first half of our system has Bad Guy #1 as the culprit. Bad Guy #1 is capable of producing large amounts of autoinducer-1, more commonly known as Acyl Homoserine Lactone. This is through constitutive expression of the luxI gene. AHL begins to diffuse into our Champion Cell, which is constitutively expressing LuxR. LuxR protein binds to the pheromone, and the AHL-LuxR complex activates the lux pR promoter, allowing the expression of green fluorescent protein and the colicin E2 activity, immunity and lysis genes (CeaB, CeiB and CelB respectively). The champion cell now fluoresces green, and colicin expression results in the lysis of the cell and the release of CeaB, which would be taken up by Bad Guy #1. Colicin E2 would lead to the eventual destruction of Bad Guy #1, and our Champion Cell will reign supreme (those that haven't burst from the production of the Colicin proteins and are still kicking around, anyway).
The first half of our system has Bad Guy #1 as the culprit. Bad Guy #1 is capable of producing large amounts of autoinducer-1, more commonly known as Acyl Homoserine Lactone. This is through constitutive expression of the luxI gene. AHL begins to diffuse into our Champion Cell, which is constitutively expressing LuxR. LuxR protein binds to the pheromone, and the AHL-LuxR complex activates the lux pR promoter, allowing the expression of green fluorescent protein and the colicin E2 activity, immunity and lysis genes (CeaB, CeiB and CelB respectively). The champion cell now fluoresces green, and colicin expression results in the lysis of the cell and the release of CeaB, which would be taken up by Bad Guy #1. Colicin E2 would lead to the eventual destruction of Bad Guy #1, and our Champion Cell will reign supreme (those that haven't burst from the production of the Colicin proteins and are still kicking around, anyway).
This system was assembled in typical iGEM fashion. The part [http://partsregistry.org/Part:BBa_J23039 J23039] was transformed into E. coli thus resulting in Bad Guy #1 cells (i.e. capable of AHL production). To construct the signaling and response circuit in the Champion Cell, [http://partsregistry.org/Part:BBa_F2620 F2620], [http://partsregistry.org/Part:BBa_I13500 I13500] and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K131009 colicin E2 operon] were assembled as shown in the picture. Colicin E2 was obtained from UC Berkeley, cloned into a TOPO vector (Invitrogen), amplified using ColicinE2 primers with flanking Biobrick Restriction Sites and cloned into a Biobrick vector.
Tests were also performed, the results of which can be seen in the Results section. These involved verifying that Bad Guy #1 is able to make AHL, verifying that our Champion Cell responds to AHL produced from Bad Guy #1, and that Colicin E2 was actually capable of preventing bacterial growth.
System #2 In Detail
![]() The other half of our system sees the aptly named Bad Guy #2 as the main adversary to our Champion Cell.
The other half of our system sees the aptly named Bad Guy #2 as the main adversary to our Champion Cell.
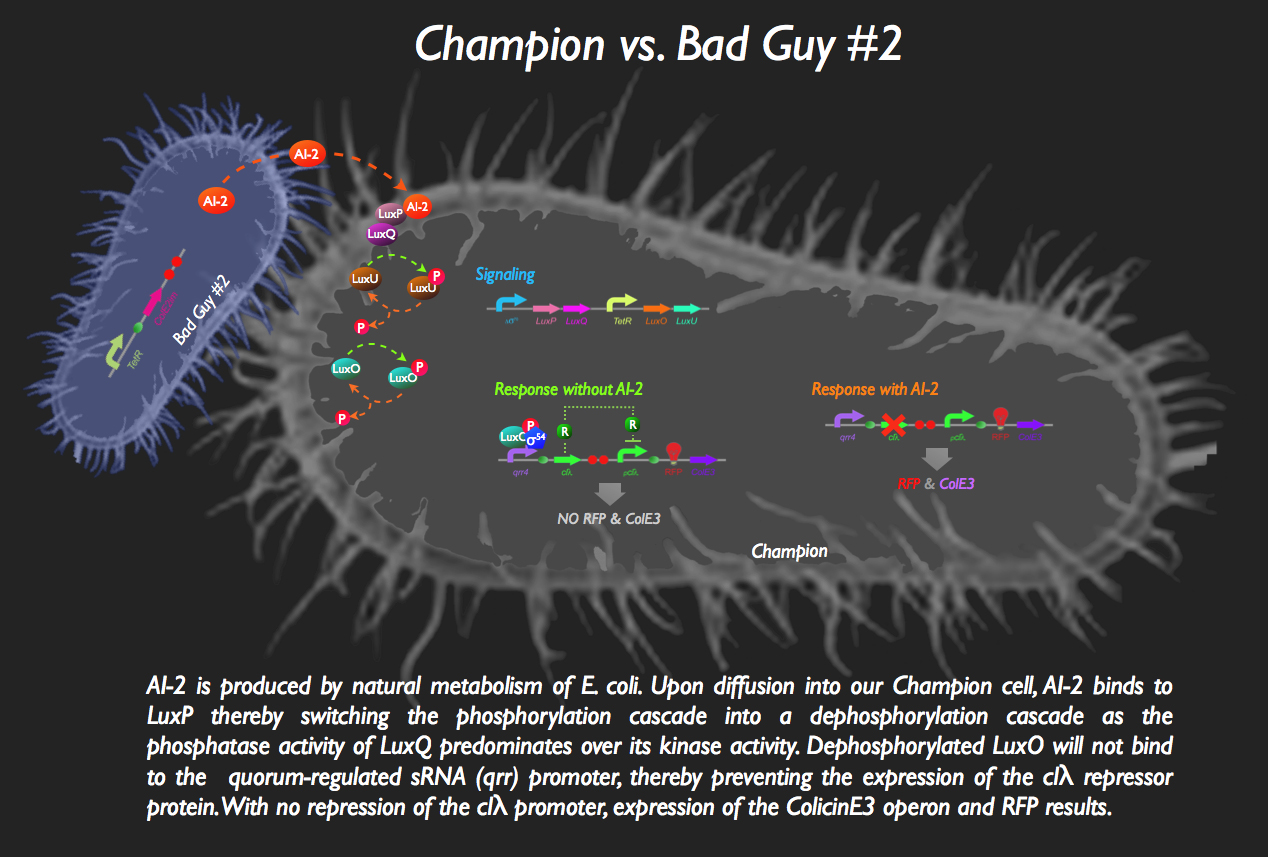
Without any AI-2 in the vicinity, however, there is downstream expression of a repressor protein, clλ, by phosphorylated LuxO, thereby masking the expression of ColicinE3 and RFP. Otherwise, without the presence of this inverter, our Champion cell would start mass production of ColE3 and RFP (indicating the presence of the villain) in the absence of any threat... the exact opposite of what we want!!
But then bad Guy #2 comes along. He’s generating AI-2 all by himself, without any engineering necessary (as a result of constitutive expression of luxS, encoding the LuxS enzyme which produces an intermediate in the AI-2 biosynthesis pathway) which is intercepted by the periplasmic protein LuxP in the Champion Cell which is constitutively bound to the LuxQ protein, a histidine kinase spanning the periplasmic membrane. The binding of AI-2 to LuxP induces the stabilization of the phosphatase conformation of the cytosolic domain of LuxQ, thereby allowing for the dephosphorylation of LuxU. The loss of phosphorylation of LuxU puts an end to its kinase activity, with the net effect being the dephosphorylation of LuxO. Since phospho-LuxO is required for the activation of the qrr4 promoter, no more clλ repressor is produced, resulting in the activation of the pclλ promoter thanks rapid destruction of the repressor afforded by the degradation tag attached to the repressor protein. ColicinE3 is transcribed, leading to the eventual destruction of Bad Guy #2, following the release of the cytotoxic protein from our champion cell. Concurrently, our Champion Cell fluoresces red to inform the viewing gallery who the invader is and in order to indicate that our champion cells are preparing to wage war!
Here's the workflow envisioned for creating system 2. Kingsley worked on the green section, Kevin was to work on the purple, Thane worked on the blue, and Dave worked on the teal.
Applications
In essence, what’s proposed above is a model or proof of concept system. Then, where could this system be useful? One evident application stems from the knowledge that specific bacterial strains create bacteriocins targeted uniquely to their populations (i.e. E. coli employ colicins while P. aeruginosa use pyocins). Combining the production of species-specific bacteriocins and reporter activity with the use of species-specific pheromone signaling systems would allow our Champion to recognize some of the real bad guys out there like Yersinia pestis (i.e., the pathogen responsible for causing Bubonic plague).
In addition, the contribution of a working of a second system for bacterial cell to cell communication (i.e., in addition to AI-1 signaling) within the Registry would drastically increase the range of future projects that could be undertaken by providing the users with an additional input (i.e. AI-2) for their genetically engineered machines.
 "
"