Team:Cambridge/Bacillus subtilis transformation
From 2008.igem.org
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{ | + | {{Cambridge/Notitle}} |
| + | <div class=bodytable> | ||
| + | {{Cambridge08}} | ||
| + | {{Cambridge08protocol}} | ||
| + | <html> | ||
| + | <table style="background:#444444; padding:15px;"> | ||
| + | <table align=left width= border="0" style="background:#444444; padding:5px;"> | ||
| + | <tr> | ||
| + | <td style="width:100%; height: 400; padding-left: 15px;"> | ||
| + | <b class="b1f"></b><b class="b2f"></b><b class="b3f"></b><b | ||
| + | class="b4f"></b> | ||
| + | <div class="contentf"> | ||
| + | <div style="height: 400; background: white; line-height:170% padding:5px;"> | ||
| + | <div style="color: white; font: 2px;">x</div> | ||
| + | </html> | ||
| + | =Transformation of ''Bacillus subtilis''= | ||
| + | ==Our aim== | ||
| - | + | We want to transform ''Bacillus subtilis'' with two kinds of vectors: episomal vectors and integration vector. We would like to obtain good efficiency, no contamination and some precise tests confirm our transformations and to be able to standardize our protocol. This protocol is modified from the one provided by the Bacillus Genetic Stock Center (http://www.bgsc.org/catalog.htm): we have made the addition of tryptophan, and experimented with different sterile methods for preparation of the growth media. | |
| - | == | + | ==Material== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
*3 different strains: | *3 different strains: | ||
| - | ** | + | **IA166 |
**IA751 | **IA751 | ||
**IA771 which contains erm insertion at the amyE locus, so transformants at the amyE locus can be screened for erythromycin resistance | **IA771 which contains erm insertion at the amyE locus, so transformants at the amyE locus can be screened for erythromycin resistance | ||
| Line 19: | Line 31: | ||
**5 shuttle vectors (3 validated) | **5 shuttle vectors (3 validated) | ||
| - | + | ==Media Preparation== | |
| - | == | + | |
''10X Medium A base:'' | ''10X Medium A base:'' | ||
| Line 47: | Line 58: | ||
* 50mM CaCl2•2H2O 0.1mL | * 50mM CaCl2•2H2O 0.1mL | ||
* 250nM MgCl2•6H2O 0.1mL | * 250nM MgCl2•6H2O 0.1mL | ||
| - | |||
''Important:'' | ''Important:'' | ||
| Line 53: | Line 63: | ||
* Store aliquots of 10X Medium A base 10mL and 10X Bacillus salts 9mL and keep them in the fridge, never use them twice to avoid contamination | * Store aliquots of 10X Medium A base 10mL and 10X Bacillus salts 9mL and keep them in the fridge, never use them twice to avoid contamination | ||
| - | == | + | ==Protocols== |
| - | === | + | ===Making Bacillus competent=== |
| - | + | # Grow one blank plate of ''Bacillus subtilis'' (or several if you want to transform different strains) for 20 hours at 37ºC (plate been kept on the bench for several days would be better) | |
| + | # Inoculate about 12mL of medium with several colonies. Mix the contents of the tube. Check with OD650. Start OD should be between 0.1 and 0.2. Be careful to pipette 0.8mL of this mixture into the cuvette to measure and dispose of it after measurement to avoid contamination in the main mixture. | ||
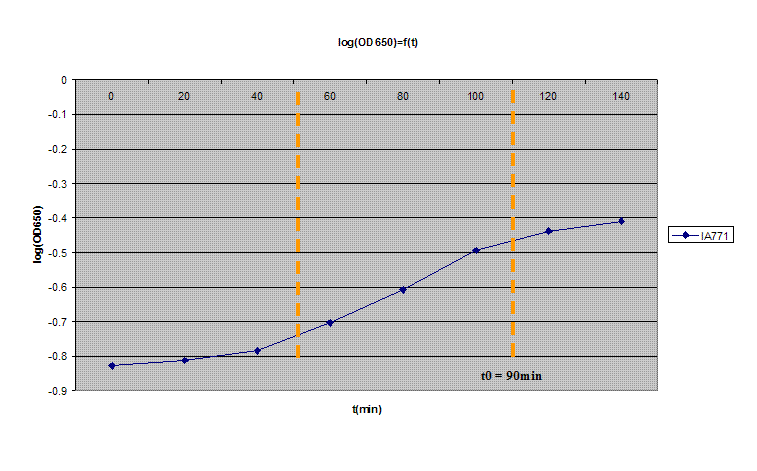
| + | # Incubate at 37ºC with vigorous shaking. Read the OD650 every 20min (never keep the solution you used for measuring!) | ||
| + | # Plot log(OD650) in function of time. After a brief lag, you should observe a exponential increase. After awhile, it will leave the exponential growth; the moment at which it leaves the exponential path is denoted as t0 (3 on the graph). It should take about 100min and the OD should be between 0.35 and 0.55. | ||
| + | [[Image:Protocol07.gif|600px|center|OD650 of ''B.subtilis'']] | ||
| + | # At t0, incubate for 90 minutes at 37ºC with vigorous shaking. | ||
| + | # Transfer 0.05mL of this culture into 0.45mL of pre-warmed Medium B in an Eppendorf tube. You have to prepare one tube for each transformation, plus an extra tube for a DNA-less control. | ||
| + | # Incubate the diluted cultures at 37ºC with shaking for 90min. At this moment, the cells are HIGHLY COMPETENT. | ||
| + | # To check for competency, you can look at cells under the microscope; competent cells are very motile. | ||
| - | + | ===Transforming=== | |
| - | + | # Spin Eppendorf tubes containing cells. Remove 400µL of liquid to keep only 100µL of the culture (to concentrate cells). Re-suspend the cell pellet in the remaining culture. | |
| + | # To transform from competent glycerol stocks, spin the tube at about 1600rpm for 20min, remove the supernatant (glycerol), and add 100µL of pre-warmed medium B. | ||
| + | # Mix the cells thoroughly. | ||
| + | # Add 0.6µg of DNA to the competent cells. | ||
| + | # Incubate for 30min at 37ºC with shaking. | ||
| + | # Plate 100µL of transformed cells onto selective agar. | ||
| - | + | ===Glycerol stocks=== | |
| - | + | ||
| - | + | ||
| - | + | # To freeze competent Bacillus cells, spin down the fresh competent cells to obtain a pellet. | |
| - | + | # Remove all supernatant. | |
| - | + | # Re-suspend cells in 500µL 60% glycerol. | |
| - | + | # Freeze tubes at -80ºC. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Results== | ==Results== | ||
| - | === | + | ===Transformation with episomal vectors=== |
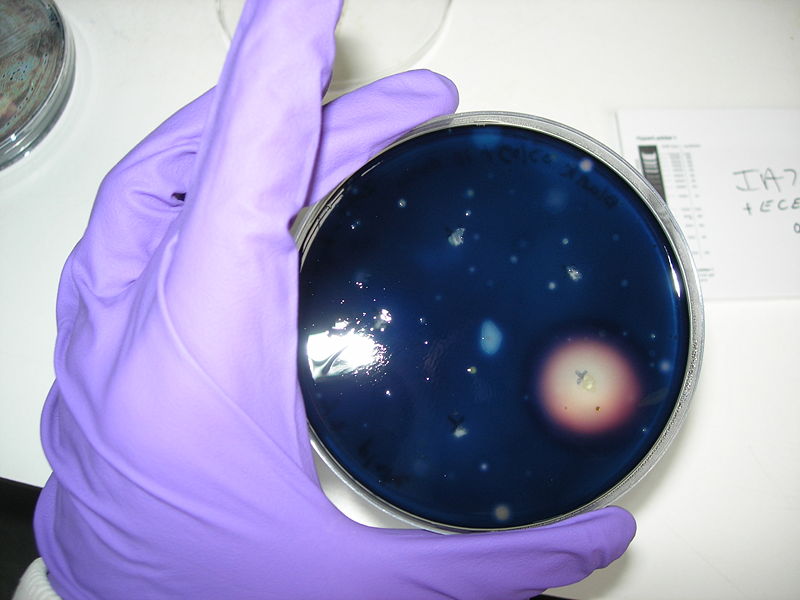
| - | ==== | + | ====ECE166==== |
* Selectable on Chloramphenicol 5µg/mL | * Selectable on Chloramphenicol 5µg/mL | ||
| Line 111: | Line 107: | ||
| - | === | + | ===Transformation with integration vectors=== |
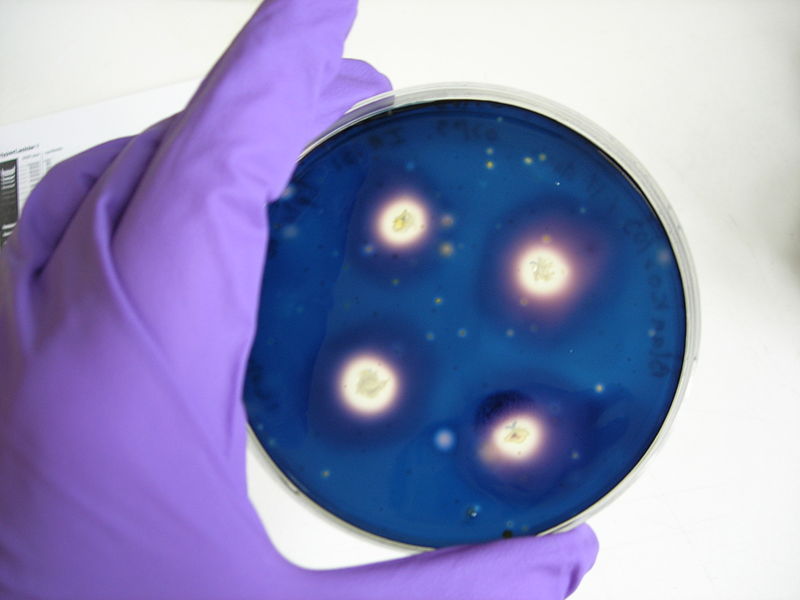
[[Image: Protocol10.jpg | thumb | 300px Positive control : IA751, big zones of clearing ]] | [[Image: Protocol10.jpg | thumb | 300px Positive control : IA751, big zones of clearing ]] | ||
[[Image: Protocol11.jpg | thumb | 300px Negative control : IA771, no zone of clearing ]] | [[Image: Protocol11.jpg | thumb | 300px Negative control : IA771, no zone of clearing ]] | ||
| - | ==== | + | ====ECE153==== |
* Selectable on Spectinomycin 50µg/mL | * Selectable on Spectinomycin 50µg/mL | ||
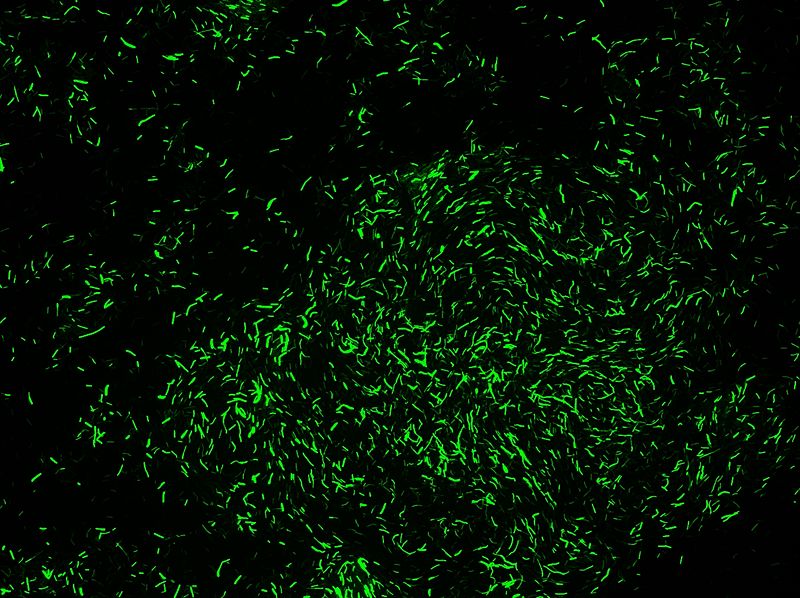
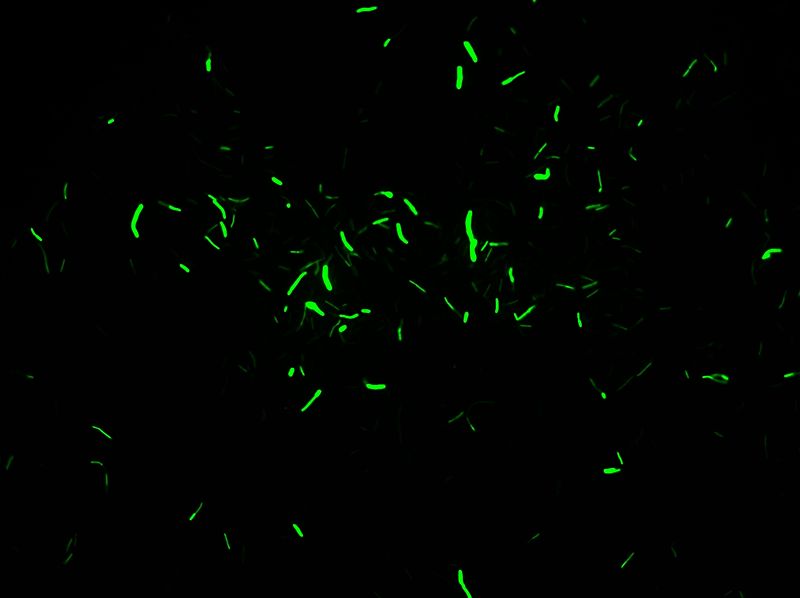
* Transformation in IA751 : Amylase test | * Transformation in IA751 : Amylase test | ||
* Xylose-inducible promoter which allow to see fluorescence | * Xylose-inducible promoter which allow to see fluorescence | ||
| - | |||
''Protocol of the Amylase Test:'' | ''Protocol of the Amylase Test:'' | ||
| Line 132: | Line 127: | ||
[[Image: Protocol12.jpg | thumb | 300px IA751 transformed with ECE153 : SUCCESSFUL TRANSFORMATION, no zone of clearing]] | [[Image: Protocol12.jpg | thumb | 300px IA751 transformed with ECE153 : SUCCESSFUL TRANSFORMATION, no zone of clearing]] | ||
| - | |||
''Protocol of the Xylose Test:'' | ''Protocol of the Xylose Test:'' | ||
| Line 143: | Line 137: | ||
We obtain good result for transformation. Amylase test is positive, but we have not manage to induce Pxyl. | We obtain good result for transformation. Amylase test is positive, but we have not manage to induce Pxyl. | ||
| - | + | ====ECE112==== | |
| - | ==== | + | |
* Selectable on Chloramphenicol 5µg/mL | * Selectable on Chloramphenicol 5µg/mL | ||
| Line 150: | Line 143: | ||
[[Image: Protocol13.jpg | thumb | 300px | center | IA751 transformed with ECE112 : SUCCESSFUL TRANSFORMATION for 4 colonies out of 5, no zone of clearing ]] | [[Image: Protocol13.jpg | thumb | 300px | center | IA751 transformed with ECE112 : SUCCESSFUL TRANSFORMATION for 4 colonies out of 5, no zone of clearing ]] | ||
| + | <html> | ||
| + | <div style="color: white; font: 2px;">x</div> | ||
| + | </div></div> | ||
| + | <b class="b4f"></b><b class="b3f"></b><b class="b2f"></b><b class="b1f"></b> | ||
| + | </td></tr></table></table> | ||
| + | </html> | ||
| - | + | __NOEDITSECTION__ | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 20:39, 29 October 2008
|
x
Transformation of Bacillus subtilisOur aimWe want to transform Bacillus subtilis with two kinds of vectors: episomal vectors and integration vector. We would like to obtain good efficiency, no contamination and some precise tests confirm our transformations and to be able to standardize our protocol. This protocol is modified from the one provided by the Bacillus Genetic Stock Center (http://www.bgsc.org/catalog.htm): we have made the addition of tryptophan, and experimented with different sterile methods for preparation of the growth media. Material
Media Preparation10X Medium A base:
10X Bacillus salts:
Medium A
Medium B
Important:
ProtocolsMaking Bacillus competent
Transforming
Glycerol stocks
ResultsTransformation with episomal vectorsECE166
We obtained consistent good results with this transformation despite the low efficiency.
Transformation with integration vectorsECE153
Protocol of the Amylase Test:
Protocol of the Xylose Test:
We obtain good result for transformation. Amylase test is positive, but we have not manage to induce Pxyl. ECE112
x
|
 "
"