Team:Cambridge/Improved GFP
From 2008.igem.org
|
x
/Testing_AHL_degradation_in_B._subtilis
IntroductionGreen Fluorescent Protein (GFP) offers efficient and convenient means of visualising the dynamic process of gene expression and of obtaining readout of the current state of complex gene regulatory networks – features of major interest for synthetic biology.
GFP and other fluorescent proteins are hence commonly used as reporter genes in synthetic biology.
Since the initial description of GFP and its potential use for molecular biology [2], many different mutant variants of this wild type have been published, conferring altered spectral properties and/or improved fluorescence intensity [5] or folding properties [4, 5] with respect to the basic protein. Improved folding and enhanced intensity of fluorescent proteins open up many new potential applications including the secretion of proteins from Gram-positive bacteria such as Bacillus subtilis, the use and efficient folding of GFP in in-vitro gene expression systems as well the potential to target markers to extracellular spaces in eukaryotic organisms such as the cell wall space in plants. In addition, more intensely fluorescing GFP variants allow the reduction in copy number of synthetic circuits carried on plasmids in bacterial hosts, thus potentially improving the stability of those systems while the reporter (GFP) is still detectable. Hence the objectives of this project were to standardise into BioBrick format, and to characterise two of the recently reported improved GFP variants in comparison to the currently used mut3GFP and to contribute those new variants to the Registry of Standard Biological Parts.
The GFP variants chosen were superfolder GFP, developed and described by Pédelacq et al (2006) [5], which was engineered for improved fluorescence in fusion proteins and P7 GFP (“superfast GFP”) which was engineered by Fisher et al (2008) [4] and selected on the basis of its property of very rapid folding.
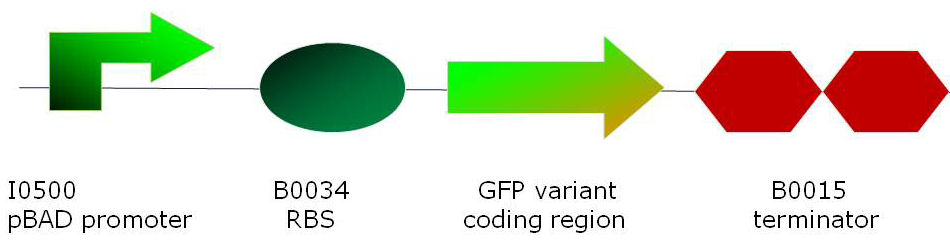
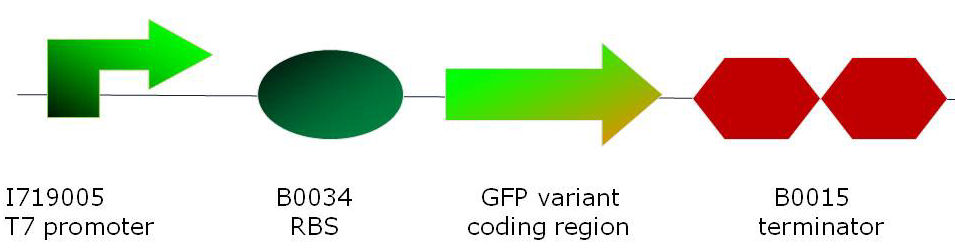
MethodsCloning of GFP variantsmut3 GFP was obtained from the 2007 parts distribution of the Registry of Standard Biological Parts [1] as part number E0400. P7 GFP [4] plasmid DNA was a kind gift by Dr Adam C. Fisher from Cornell University, New York. Superfolder GFP [5] was created by de novo DNA synthesis (Geneart) and its codon bias was changed as a compromise for optimum expression in E. coli and B. subtilis. The full sequence is listed in the Registry of Standard Biological parts: part number <a href="http://partsregistry.org/Part:BBa_I746916">I746916</a>.
For in vivo and in vitro studies, all three GFP variants (superfolder, P7 and mut3) were cloned into 2 basic constructs residing in the same standard vector (pSB1A2):
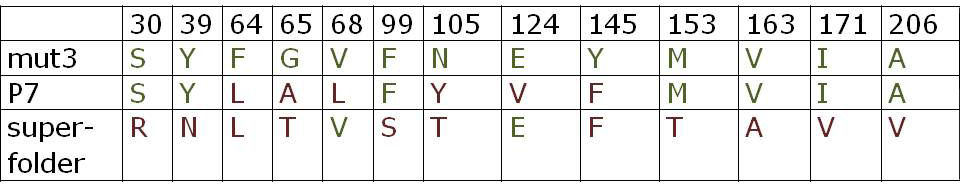
All constructs were sequenced after assembly to confirm sequence identity with the GFP variants in the original papers [2,3]. Below is an overview of the mutations of P7 GFP and superfolder GFP in comparison to mut3 GFP:
In vivo fluorescence analysisOvernight cultures of E.coli BW27783 carrying the one of the GFP constructs (see table above) on pSB1A2 were diluted 1:10 into fresh LB with 100ug/ml ampicillin and grown in a 96-well plate at 37°C with shaking for 2 hours before induction with varying concentrations of L-arabinose. Fluorescence was measured throughout the duration of the experiment in 3minute intervals on a BMG FluoStar fluorescence plate reader with 485nm excitation, 520nm emission filters.In vitro folding and spectral analysis6-his tagged GFP variants were purified using hybrid purification conditions in the ProBond purification system (Invitrogen). Peak elution fractions were identified by analysis on 12% SDS PAGE. For folding analysis purified GFP variants were diluted 1:10 into TNG buffer (25mM Tris pH 7.5, 0.2M NaCl, 5% v/v glycerol) with 6.8M guanidine hydrochloride and heated to 95°C for 5 minutes. Following this denaturation step, samples were dispensed into a 96 well plate and a single reading of GFP fluorescence intensity was taken from each sample. Samples were then diluted 1:10 into fresh TNG buffer containing no guanidine hydrochloride and fluorescence recovery was measured in 15 second intervals immediately after dilution. Purified samples diluted 1:10 into TNG without guanidine hydrochloride for the baseline read and again 1:10 into fresh TNG for further readings served as positive (native, non-denatured) controls while samples denatured as described above and diluted 1:10 into TNG with 6.8M guanidine hydrochloride were negative (fully unfolded) controls.ResultsSuperfolder GFP appears much brighter to the oberserver than P7 or mut3 GFP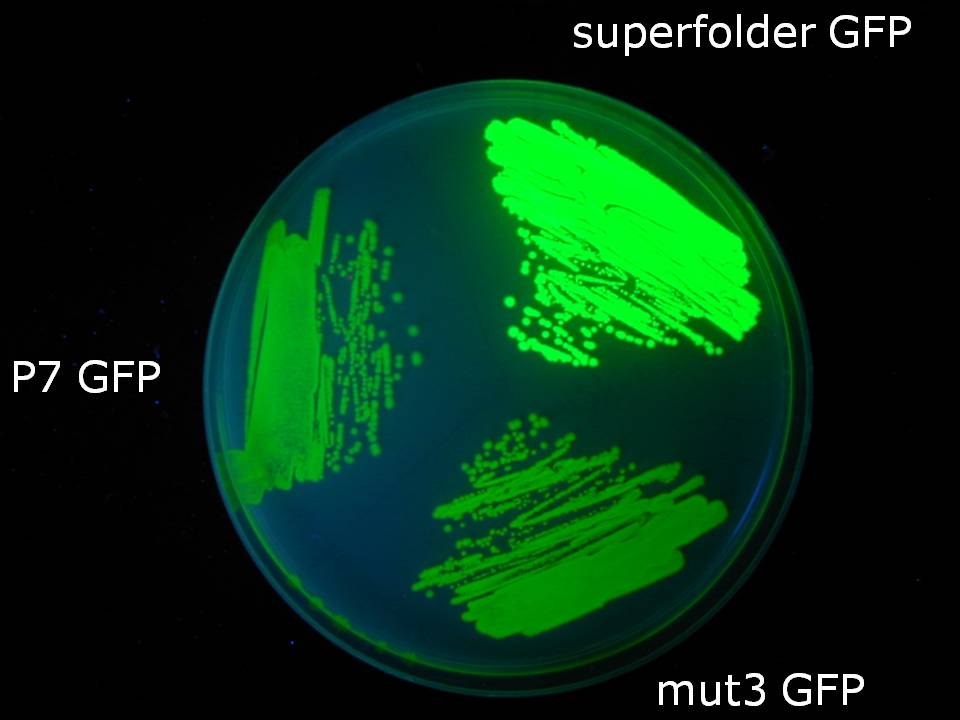
Figure 0: E.coli BW27783 growing on LB agar plate containing 10mM arabinose. The three streaks are expressing the GFP variant indicated in the picture driven by the pBAD promoter (see Methods for construct). Plate was illuminated with handheld UV lamp and photographed. Superfolder GFP fluorescence increases 3.5 to 4 fold faster than P7 or mut3 GFP fluorescence in vivo: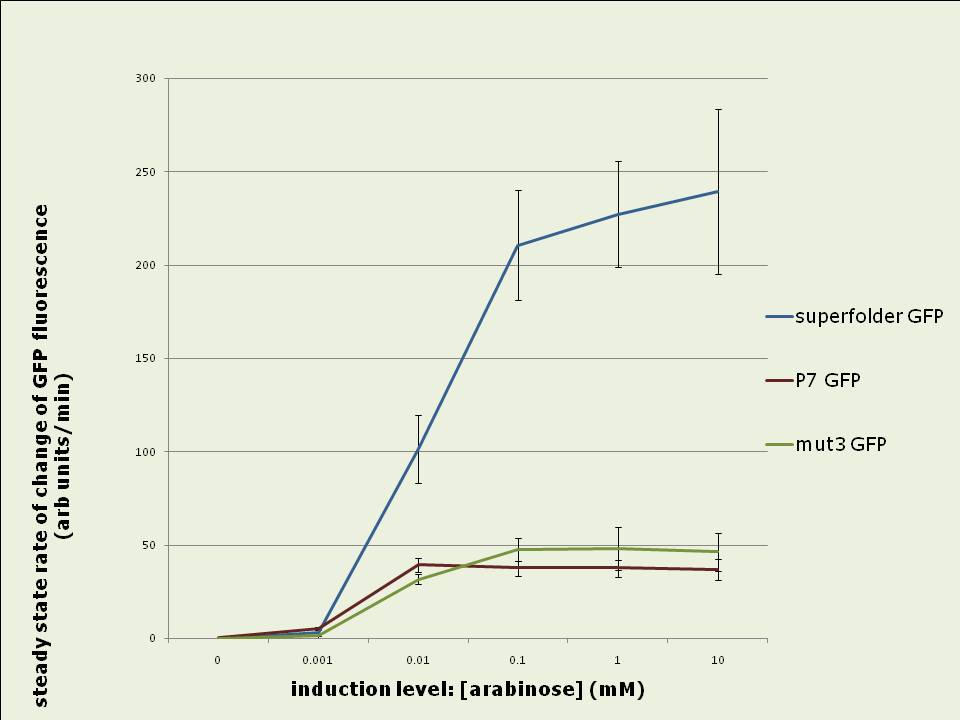
Figure 1: Rates of increase of in vivo GFP fluorescence 2.5 to 4hrs after induction of the pBAD promoter with different levels of arabinose. Data shown is the average of 3 experiments. Superfolder GFP yields up to four fold more total fluorescence intensity than mut3 or P7 GFP when expressed in vivo: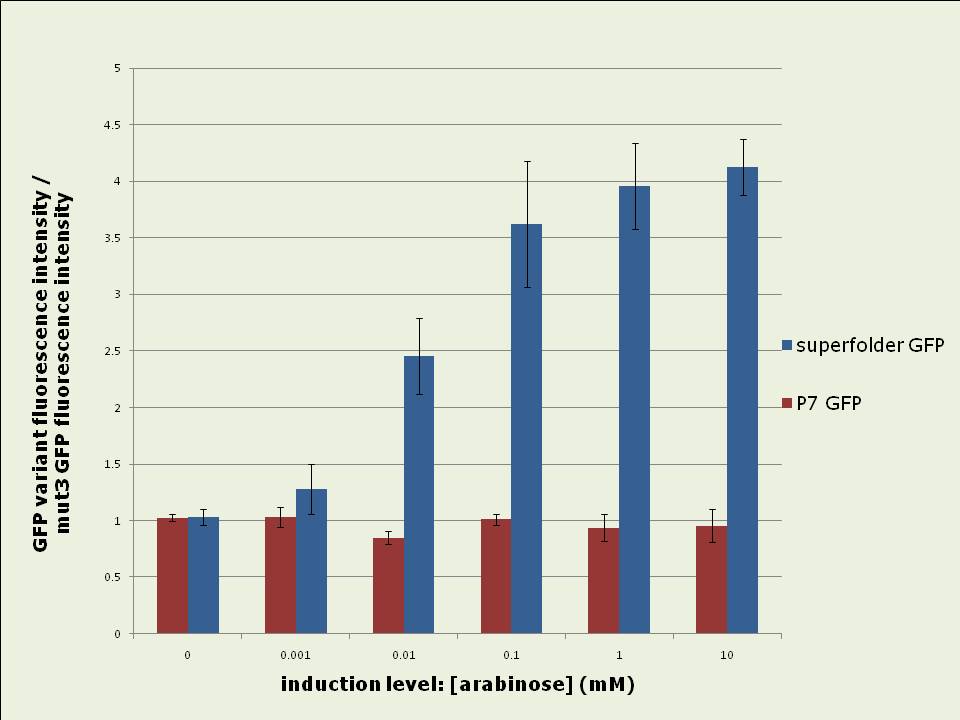
Figure 2: In vivo fluorescence intensities of GFP variants were compared at steady state (between 3hrs – 5hrs after induction). The fluorescence intensities of superfolder and P7 GFP are expressed as fractions of the intensity of mut3 GFP. The data shown is the average of 3 experiments. Excitation and emission spectra vary slightly between the GFP variants: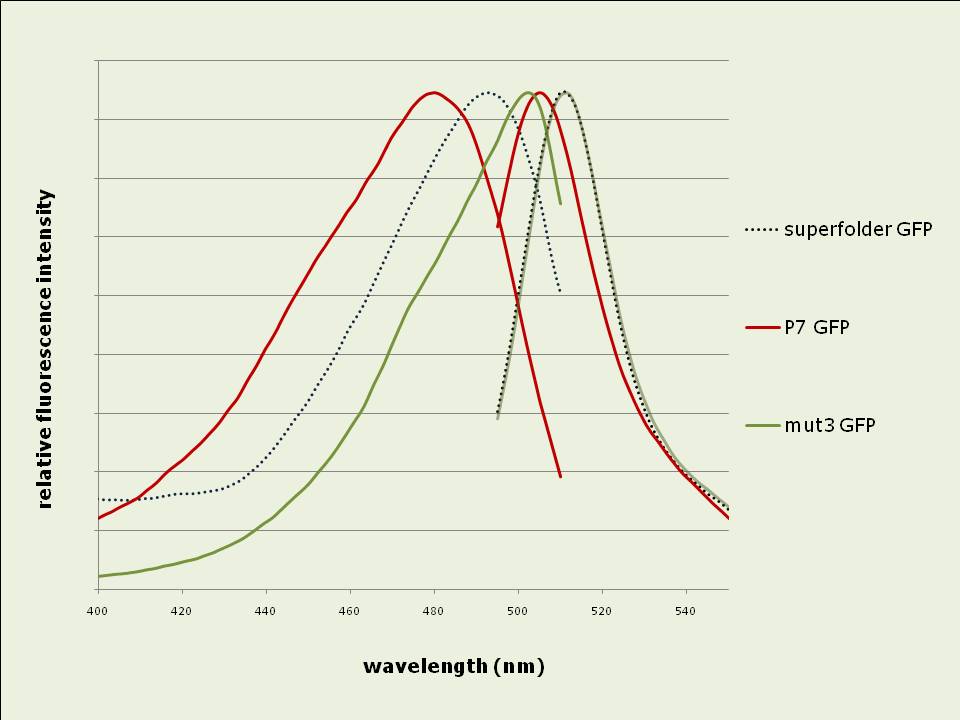
Figure 3: Excitation and Emission spectra determined for 6-his purified GFP variants. Excitation was at 480nm for emission spectra; emission was at 520nm for excitation spectra. Emission peaks occur at 479, 492 and 502nm while excitation peaks are located at 505, 511 and 511nm for P7, superfolder and mut3 GFPs respectively. After denaturing in vitro, P7 GFP renatures faster than superfolder or mut3 GFP: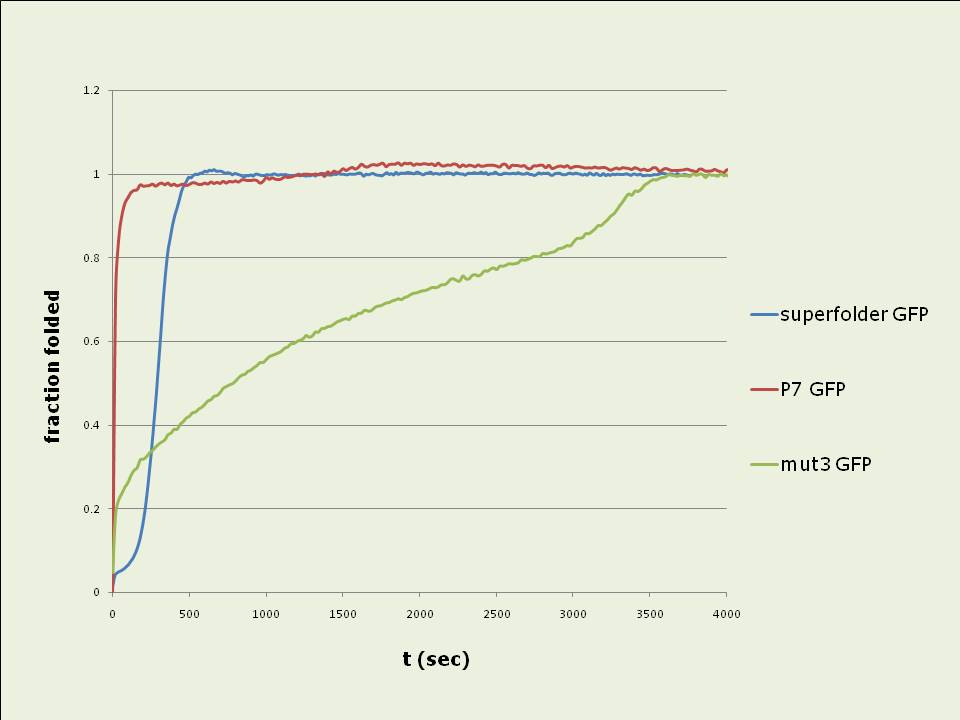
Figure 4: Short term in vitro refolding of 6-his purified GFP samples after complete denaturing. Data shown is the fraction folded at each time point compared to the maximally refolded sample. Data shown is the average of three experiments. P7 and superfolder GFPs are more resistant to denaturing and have stronger refolding abilities in vitro than mut3 GFP: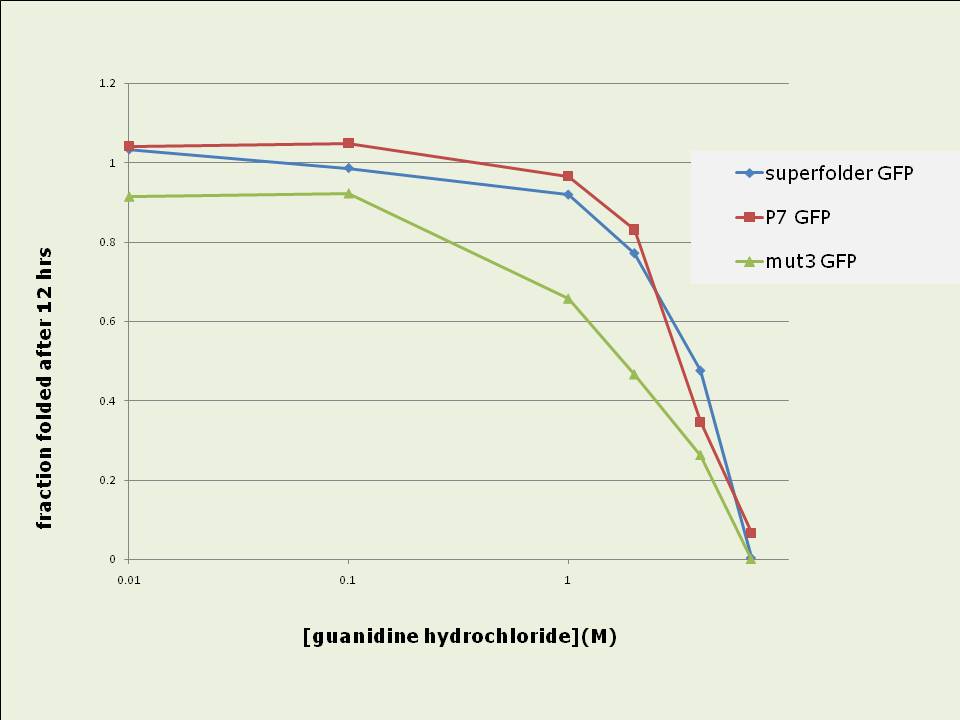 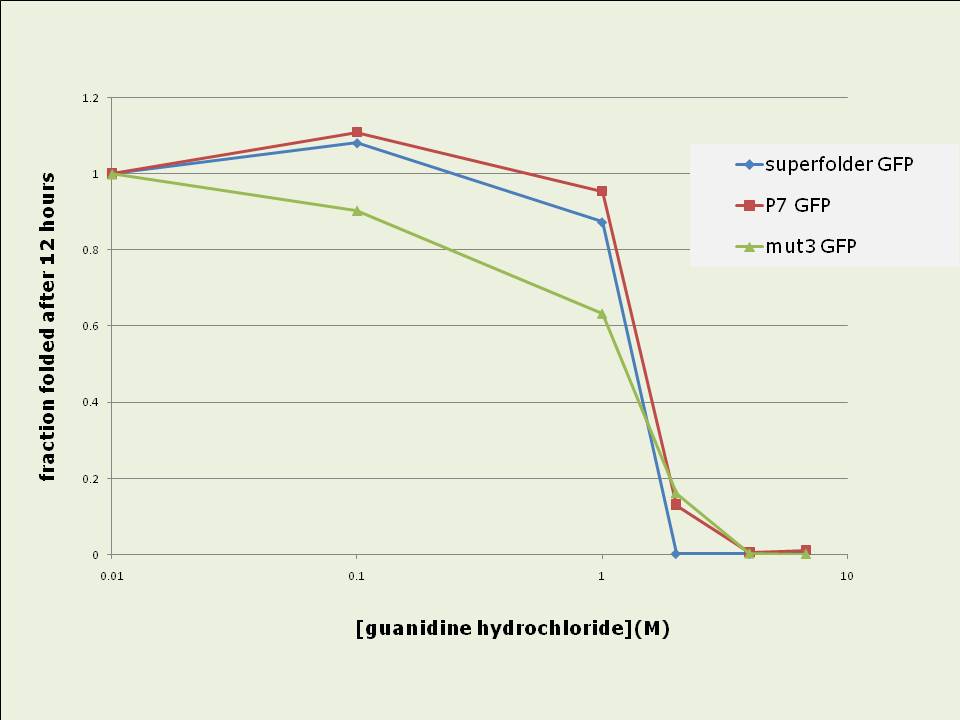
Figure 5 (left): Resistance of 6-his purified GFP variants to denaturing after 100fold dilution into TNG buffer containing different concentrations guanidine hydrochloride Data shown is the fraction folded after 12 hours compared to a fully folded sample. Figure 6 (right): Ability of 6-his purified GFP variants to refold in different concentrations guanidine hydrochloride after complete denaturation. Data shown is the fraction folded after 12 hours compared to a fully folded sample. Data shown for both cases are averages of three experiments. ConclusionTwo improved GFP variants were standardised into BioBrick format and their in vivo and in vitro properties were compared to mut3 GFP:Superfolder GFP represents a major improvement for in vivo GFP expression and will hence be extremely useful for synthetic biology applications: Its 4fold increased fluorescence intensity and the 3.5 fold higher speed at which it develops fluorescence in vivo are likely to be very useful features, especially with regard to the reduction of the copy number of synthetic regulatory circuits to low- or single-copy whilst still maintaining the ability to use and detect GFP as a reporter. P7 GFP, whose in vivo properties are essentially equal to those of mut3 GFP, may be a promising and useful GFP candidate for in vitro experiments, given its properties of very fast in vitro refolding, its high resistance to denaturing and its improved ability to renature . Both new variants were contributed to the Registry of Standard Biological Parts and are as such available for wide use in the synthetic biology community. References[1] MIT registry of standard biological parts: http://partsregistry.org[2] Chalfie, et al. (1994). "Green Fluorescent Protein as a Marker for Gene Expression," Science 263, 802-805. [3] Cormack et al (1996) “FACS-optimized mutants of the green fluorescent protein (GFP)”, Gene, 173, 33-38 [4] Fisher et al (2008) “Laboratory Evolution of Fast-Folding Green Fluorescent Protein Using Secretory Pathway Quality Control”, PLoS ONE 3(6) [5] Pédelacq et al (2006) “Engineering and characterization of a superfolder green fluorescent protein”, Nature Biotech 24 (1), 79-88 Links[1] PDF of a poster created to present the above data to a mixed audience. Includes an introductory section on BioBricks. AcknowledgementsThis undergraduate summer research project was supported by the Wellcome Trust in form of a vacation scholarship. It was conducted in the lab of Dr Jim Haseloff, Dept of Plant Sciences, University of Cambridge, UK. We wish to thank Dr Adam C. Fisher from Cornell University, New York for his kind donation of P7 GFP. x
|
 "
"