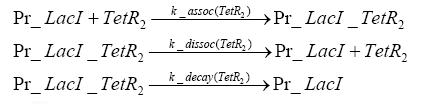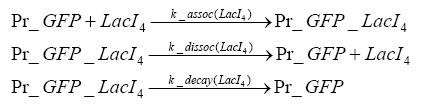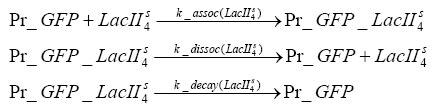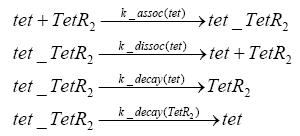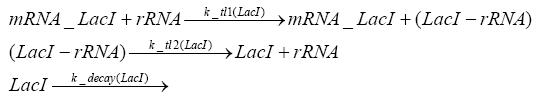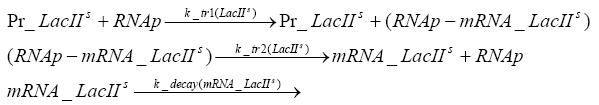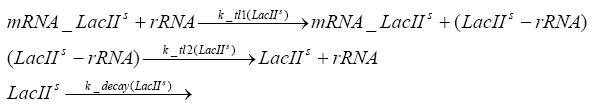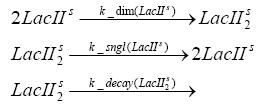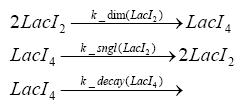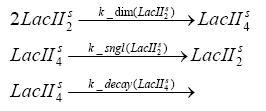Team:ETH Zurich/Modeling/Switch Circuit
From 2008.igem.org
(Difference between revisions)
(→References) |
(→References) |
||
| Line 182: | Line 182: | ||
(2) ''"Predicting stochastic gene expression dynamics in single cells"'', Mettetal et al., ''PNAS'', 2006 | (2) ''"Predicting stochastic gene expression dynamics in single cells"'', Mettetal et al., ''PNAS'', 2006 | ||
| + | (3) ''"Engineered gene circuits"'', Hasty et al., ''Nature'', 2002 | ||
<!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | <!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | ||
|} | |} | ||
Revision as of 18:15, 27 October 2008
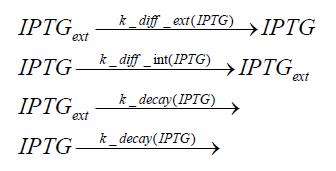
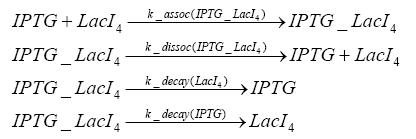
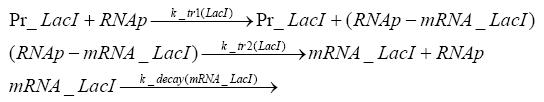
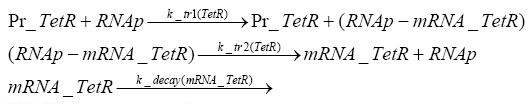
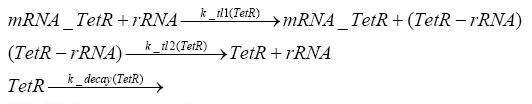
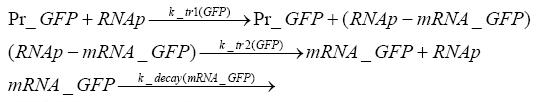
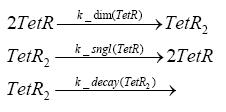
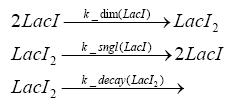
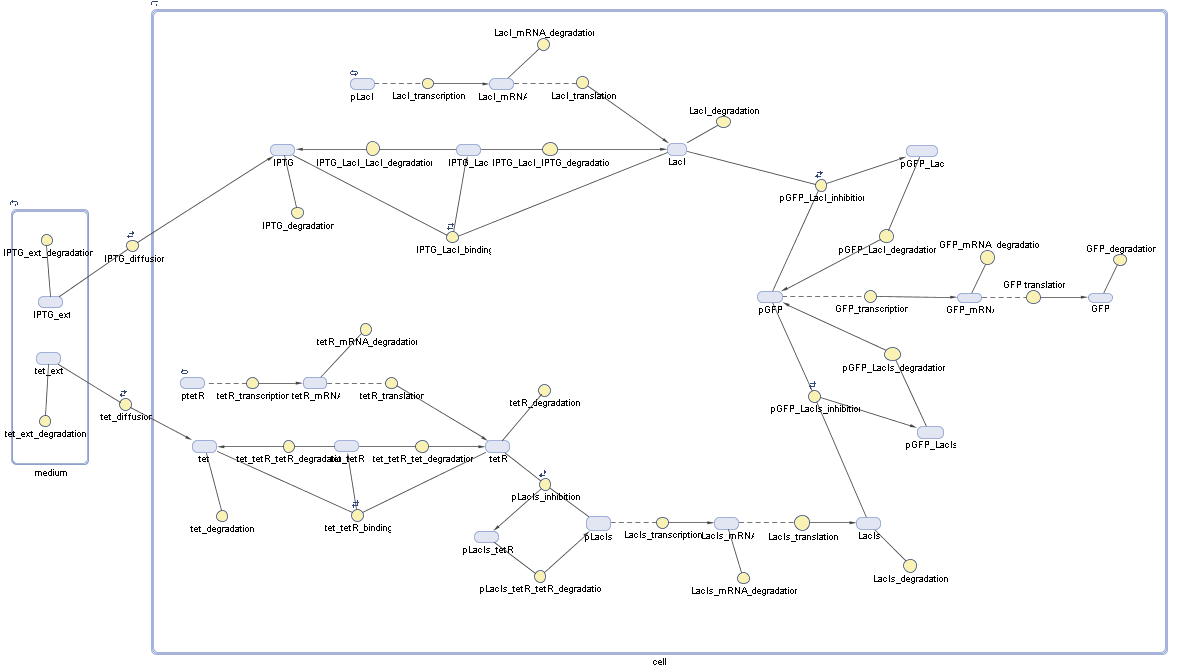
Switch CircuitDetailed Modeldiffusion of IPTGdiffusion of tetbinding of tetR to LacI-promotor and LacIIs-promotorbinding of LacI and LacIIs to GFP-promotorbinding of tet to tetRbinding of IPTG to LacItranscription and translation of LacItranscription and translation of LacIIstranscription and translation of tetRtranscription and translation of GFPdimerization of tetRdimerization and tetramerization of LacI and LacIIsImplementationThe model of the switch circuit has been implemented using the Simbiology Toolbox in MATLAB. For the sake of simplicity, the effects of dimerization and dimerization/tetramerization of tetR and LacI/LacIIs have been neglected. So we did with the additional steps involving the RNApolymerase in the transcription of proteins and the ribosomes in the translation of proteins. SimulationResults and DiscussionParametersIn this section you can find all the parameters used in the simulation.
References(1) "Spatiotemporal control of gene expression with pulse-generating networks", Basu et al., PNAS, 2004 (2) "Predicting stochastic gene expression dynamics in single cells", Mettetal et al., PNAS, 2006 (3) "Engineered gene circuits", Hasty et al., Nature, 2002 |
 "
"