Team:ETH Zurich/Project/Overview
From 2008.igem.org
Luca.Gerosa (Talk | contribs) (→Overview: The Minimal Genome Project) |
|||
| (4 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
| - | The reduction of a genome to its most fundamental parts is a task of broad biological interest which has | + | The reduction of a genome to its most fundamental parts is a task of broad biological interest which has not been achieved yet. It addresses the question of which set of genes is absolutely necessary to support life in its most primitive form. |
Several approaches have been taken by a number of scientists to tackle this problem. Recently, a cleaned-up version of ''E. coli'' containing a 15% smaller genome has been presented by Posfai et al. In this approach, however, targeted deletion of genes was applied. This requires an informed, rational step-by-step approach. Therefore, it cannot apply one of the features of biology which allow to explore a much wider space with frequently unexpected outcomes: evolution. | Several approaches have been taken by a number of scientists to tackle this problem. Recently, a cleaned-up version of ''E. coli'' containing a 15% smaller genome has been presented by Posfai et al. In this approach, however, targeted deletion of genes was applied. This requires an informed, rational step-by-step approach. Therefore, it cannot apply one of the features of biology which allow to explore a much wider space with frequently unexpected outcomes: evolution. | ||
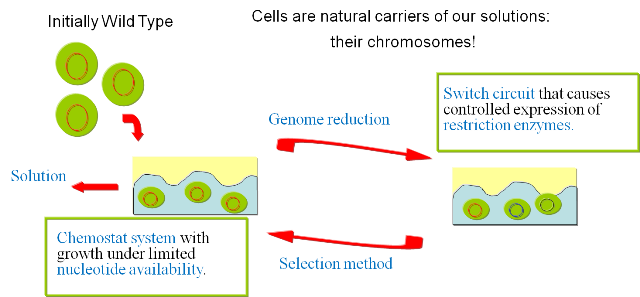
| - | + | We here propose a novel approach that is based on the concept of evolutionary algorithm, borrowed from Computer Science and it is implemented in a (synthetic) biology scenario. Starting from the observation that solving the minimal genome problem is both a question of huge solution space (too big to be even computed on a pc) and of practical nature (how to obtain it finally in our hands?), we here propose an approach that aim to solve at once both problematics. Starting from the concept that cells are natural carriers of our solutions (they have a chromosome), we design a mechanism in which our solutions (chromosomes) are mutated (reduced) and only the fitter are brought to the next solution population. In this way cells are both solution carriers and concrete results to our problem. | |
| + | |||
| + | Genome reduction is achieved by short pulses of two ''in vivo'' synthesized enzymes, a restriction enzyme and a ligase. Synthetic Biology concepts are used to construct the needed control genetic circuit. The restriction enzyme randomly cuts the genomic DNA. If two cuts or more are generated within one pulse, a chromosomal fragment might be excluded before the ligase performs its job of religation. This results in the random deletion of chromosomal fragments. | ||
Though the chance of this event to happen is rather low considering a single cell, it becomes likely when looking at an entire population. However, this creates the need of having to select for cells which have successfully eliminated parts of their genome. Since manual selection is obviously impossible considering the tremendous number of cells in a culture, it would be convenient to equip cells carrying a small genome with some kind of growth advantage enabling them to outgrow the rest of the population in a continuous culture. | Though the chance of this event to happen is rather low considering a single cell, it becomes likely when looking at an entire population. However, this creates the need of having to select for cells which have successfully eliminated parts of their genome. Since manual selection is obviously impossible considering the tremendous number of cells in a culture, it would be convenient to equip cells carrying a small genome with some kind of growth advantage enabling them to outgrow the rest of the population in a continuous culture. | ||
| Line 27: | Line 29: | ||
Therefore, we opted for putting a constraint on nucleotide synthesis to make DNA replication the rate-limiting step of proliferation. In this case, cells with a smaller genome should replicate and therewith divide faster than cells carrying larger genomes. | Therefore, we opted for putting a constraint on nucleotide synthesis to make DNA replication the rate-limiting step of proliferation. In this case, cells with a smaller genome should replicate and therewith divide faster than cells carrying larger genomes. | ||
| - | Finally, after the last pulse has been applied, the ''E. coli'' carrying | + | Finally, after the last pulse has been applied, the ''E. coli'' carrying a dramatically reduced genome will take the lead in our culture. If the nucleotide availability constraint turns indeed out to be a powerful selection mechanism, this method might take us a long way to obtaining a reduced genome. Our approach of creating an ''E. coli'' showing a minimal genome would be thus largely free of any bias imprinted by our current understanding of E. coli physiology (except, of course, for the applied selection criteria). |
| + | See [[Team:ETH Zurich/Project/Motivation|Motivation page]] for details. | ||
| + | |||
<!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | <!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | ||
|} | |} | ||
Latest revision as of 01:08, 30 October 2008
|
Overview: The Minimal Genome ProjectThe reduction of a genome to its most fundamental parts is a task of broad biological interest which has not been achieved yet. It addresses the question of which set of genes is absolutely necessary to support life in its most primitive form. Several approaches have been taken by a number of scientists to tackle this problem. Recently, a cleaned-up version of E. coli containing a 15% smaller genome has been presented by Posfai et al. In this approach, however, targeted deletion of genes was applied. This requires an informed, rational step-by-step approach. Therefore, it cannot apply one of the features of biology which allow to explore a much wider space with frequently unexpected outcomes: evolution. We here propose a novel approach that is based on the concept of evolutionary algorithm, borrowed from Computer Science and it is implemented in a (synthetic) biology scenario. Starting from the observation that solving the minimal genome problem is both a question of huge solution space (too big to be even computed on a pc) and of practical nature (how to obtain it finally in our hands?), we here propose an approach that aim to solve at once both problematics. Starting from the concept that cells are natural carriers of our solutions (they have a chromosome), we design a mechanism in which our solutions (chromosomes) are mutated (reduced) and only the fitter are brought to the next solution population. In this way cells are both solution carriers and concrete results to our problem. Genome reduction is achieved by short pulses of two in vivo synthesized enzymes, a restriction enzyme and a ligase. Synthetic Biology concepts are used to construct the needed control genetic circuit. The restriction enzyme randomly cuts the genomic DNA. If two cuts or more are generated within one pulse, a chromosomal fragment might be excluded before the ligase performs its job of religation. This results in the random deletion of chromosomal fragments. Though the chance of this event to happen is rather low considering a single cell, it becomes likely when looking at an entire population. However, this creates the need of having to select for cells which have successfully eliminated parts of their genome. Since manual selection is obviously impossible considering the tremendous number of cells in a culture, it would be convenient to equip cells carrying a small genome with some kind of growth advantage enabling them to outgrow the rest of the population in a continuous culture.
Therefore, we opted for putting a constraint on nucleotide synthesis to make DNA replication the rate-limiting step of proliferation. In this case, cells with a smaller genome should replicate and therewith divide faster than cells carrying larger genomes. Finally, after the last pulse has been applied, the E. coli carrying a dramatically reduced genome will take the lead in our culture. If the nucleotide availability constraint turns indeed out to be a powerful selection mechanism, this method might take us a long way to obtaining a reduced genome. Our approach of creating an E. coli showing a minimal genome would be thus largely free of any bias imprinted by our current understanding of E. coli physiology (except, of course, for the applied selection criteria). See Motivation page for details. |
 "
"