Team:Edinburgh/Plan
From 2008.igem.org
Contents |
An ambitious plan
The chassis: E. coli vs. B. subtilis
We were initially torn between using Escherichia coli or Bacillus subtilis as a chassis for development of MicroMaize:
We decided that B. subtilis would have advantages in the case of our system working and industrialisation being an option. This was because of increased ease of transport (due to its stable spore-forming nature), its having a better characterised secretory system and being regarded in higher favour than E. coli by the general public (see section of ethical, legal and social implications). Another possibility for the future would be to port the system to yeast, Saccharomyces cerevisiae; this would be more difficult to achieve but might ultimately be more desirable as yeast is Generally Regarded As Safe for food use, and as a eukaryote, might be able to accumulate larger amounts of starch than bacteria.
However, for the labwork we decided to use E. coli JM109 as the chassis. This was due to the greater availability of tested registry parts for E. coli compared to B. subtilis, a higher level of understanding of the biochemistry of E. coli, and more in-house experience of engineering E. coli.
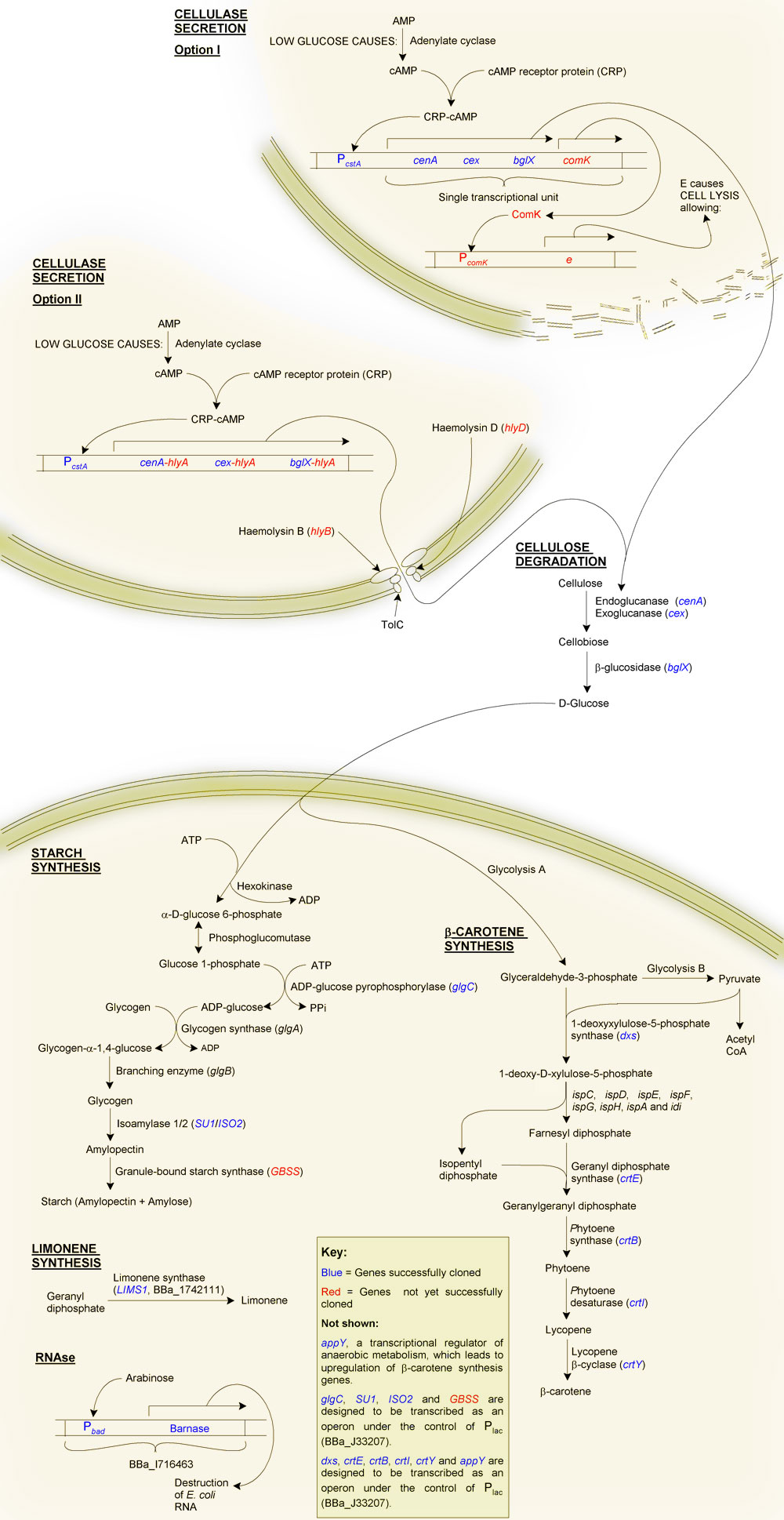
Primary Objective: The production of starch from cellulose
Firstly, why starch?
Although one of our aims is to provide a substrate for the biofuels industry, which can easily ferment glucose to ethanol, we decided to try to synthesise starch instead, even though this requires extra steps. The reason for this was a question of purification: glucose is soluble, and therefore difficult to purify from solution, as is glycogen. Starch on the other hand is highly insoluble making purification of this end product relatively straight forward. As well as this, glucose becomes inhibitory at high concentrations due to osmotic pressure, and may also cause product inhibition of glucose-producing reactions. Starch, being insoluble, can potentially accumulate at much higher quantities without disrupting the biochemistry of the cell.
The production of starch from cellulose requires two main steps:
Secondary Objective: The synthesis of β-carotene
β-carotene is the pigment that gives carrots their orange colour. It is produced as an intermediate of the lycopene synthesis pathway and is converted into vitamin A by the human body. We added a number of genes from Pantoea ananatis (formerly Erwinia ananas) to E. coli, in essence transferring the beta-carotene synthesis pathway of P. ananatis to our chasis organism. We also cloned two E. coli genes in order to upregulate the transgenic β-carotene synthesis pathway.
Further considerations
As well as designing the major components of our system, we had to consider how we would make MicroMaize a safe and appealling product (see ELSI page for more details).
An inducible RNAse
Although we can easilly kill E. coli through heat treatment, the cells contain a lot of RNA, which can cause gout. To overcome this we decided to use the "genetic self-destruct" system (BBa_I716463) developed by UC Berkley's '07 team.
Briefly, this is an arabinose-inducible RNAse. Its use for us is that we can grow our cells to the required starch or β-carotene content, then induce transcription of the RNAse Barnase. Barnase will then hydrolyse the RNA into single nucleotides. We envisage that these will then diffuse through the bacterial cell wall after disruption through heat treatment.
Lemon flavour
System overview
References
Cellulose degradation
- Lynd, L.R., Weimer, P.J., van Zyl, W.H., and Pretorius, I.S. 2002. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiology and Molecular Biology Reviews 66, 506-577.
Glycogen overproduction
- Leung, P., Lee, Y.M., Greenberg, E., Esch, K., Boylan, S., and Preiss, J. 1986. Cloning and expression of the Escherichia coli glgC gene from a mutant containing an ADP-glucose pyrophsophorylase with altered allosteric properties. Journal of Bacteriology 167, 82-88.
- Eydallin, G., Viale, A.M., Moran-Zorzano, M.T., Munoz, F.J., Montero, M., Baroja-Fernandez, E., and Pozueta-Romero, J. 2007. Genome-wide screening of genes affecting glycogen metabolism in Escherichia coli K-12. FEBS Letters 581, 2947-2953.
- Dedhia, N., Chen, W., and Bailey, J.E. 1996. Design of expression systems for metabolic engineering: coordinated synthesis and degradation of glycogen. Biotechnology and Bioengineering 55, 419-426.
- Fernandez-Banares, I., Clotet, J., Arino, J., and Guinovart, J.J. 1991. Glycogen hyperaccumulation in Saccharomyces cerevisiae RAS2 mutant - a biochemical study. FEBS Letters 290, 38-42.
Starch biosynthesis
- Ball, S.G. and Morell, M.K. 2003. From bacterial glyogen to starch: understanding the biogenesis of the plant starch granule. Annual Reviews in Plant Biology 54, 207-233. (A comprehensive review of the state of knowledge regarding glycogen and starch biosynthesis in bacteria and plants).
- Delatte T et al. 2005. Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant Journal 41, 815-830. (Probable role of the heteromeric isoamylase in converting a glycogen-like precursor to starch).
- Wattebled F et al. 2005. Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiology 138, 184-195. (Probable role of the heteromeric isoamylase in converting a glycogen-like precursor to starch).
- Kubo A et al. 2005. Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 in gene in supports a direct role for isoamylase1 amylopectin biosynthesis. Plant Physiology 137, 45-36. (Probable role of the heteromeric isoamylase in converting a glycogen-like precursor to starch).
- Utsumi, Y., and Nakamura, Y. 2006. Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the isoamylase1-isoamylase2 hetero-oligomer from rice endosperm. Planta 225, 75-87. (Expression of functional isoamylase subunits in E. coli shows that this enzyme can be expressed in a functional form in bacteria).
Synthesis of carotenoids
- Sandmann, G. 2006. Production of carotenoids by gene combination in Escherichia coli. pages 143-153 in 'Food Biotechnology', 2nd edition, eds. K. Shetty, G. Paliyath, A. Pometto and R.E. Levin, CRC Press, Taylor & Francis group, Boca Raton, FL. (A useful review).
- Misawa, N., Nakagawa, N., Kobayashi, K., Yamano, S., Nakamura, K., and Harashima, K. 1990. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. Journal of Bacteriology 172, 6704-612. (Demonstration of the functional expression of the Pantoea ananatis (= Erwinia uredovora) genes in E. coli)
- Kang, M.J., Lee, Y.M., Yoon, S.H., Kim, J.H., Ock, S.W., Jung, K.H., Shin, Y.C., Keasling, J.D., and Kim, S.W. 2005. Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. biotechnology and Bioengineering 91, 636-642 (role of dxs and appY in increasing carotenoid biosynthesis).
Synthesis of limonene and other terpenes
- Lucker, J., El Tamer, M.K., Schwab, W., Verstappen, F.W.A., van der Plas, L.H.W., Bouwmeester, H.J., and Verhoeven, H.H. 2002. Monoterpene biosynthesis in lemon (Citrus limon). European Journal of Biochemistry 269, 3160-3171. (Functional expression of the LIMS1 gene in E. coli).
- Reiling, K.K., Yoshikuni, Y., Martin, V.J.J., Newman, J., Bohlmann, J., and Keasling, J.D. 2004. Mono and diterpene production in Escherichia coli. Biotechnology and Bioengineering 87, 200-212. (Generation of various monoterpenes, including limonene, in E. coli and methods for detection).
 "
"