Team:Harvard/Dailybook/Week10/Chemical and Light
From 2008.igem.org
Contents |
Transformations in Shewie
About 250 ng of DNA added each time.
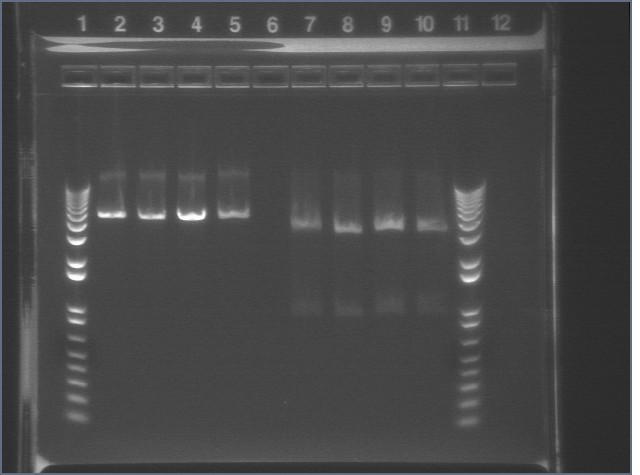
First Set 8/22
| Plate | Colonies | Size | Color |
| S1 P102 (P108+45) Kan | 5 colonies | 3mm to 5 mm | Pink |
| S1 P121+P38 (P124) 1:2.5 Kan A | 1 colony | 5mm to 7.5mm | Pink |
| S1 P28 (ColE1) Kan | 0 colonies | ||
| S1 P121+P38(P124) 1:2.5 Kan B | 0 colonies | ||
| S1 P121+P39 1:2.5 Dephos Kan A | 0 colonies | ||
| S1 Topo vector (puc) 2ul Kan X-gal | >50 colonies | 1mm | White |
| S1 Topo vector (puc) 4ul Kan X-gal | >50 colonies | 1mm | White |
| S1 P121+P39 1:2.5 Kan D | 0 colonies | ||
| S1 (-) ctrl (just cells) Kan | 0 colonies | ||
| S1 (+) ctrl (E1 p59b) Kan | 2 colonies | 5mm | Pink |
Poor transformation efficiency in the positive control suggests that there may be a problem with our cells. All samples, except for P102, were retransformed the following week with fresh cell cultures.
Second Set (with new cells) 8/27
| Plate | Marker | Description |
| S1 P124 1:2.5 A | Kan | 14 2-3mm pink colonies, ~100 0.5mm white colonies |
| S1 P124 1:2.5 B | Kan | 2 pink 2mm colonies, >100 0.5mm white colonies |
| S1 P125 1:2.5 Dephos A | Kan | 2 pink 2mm colonies, >100 0.5mm white colonies |
| S1 P125 1:2.5 D | Kan | 8 2mm pink colonies, ~100 0.5mm white colonies |
| S1 P28 | Kan | ~8 1-2mm pink colonies, ~50 0.5mm white colonies |
| S1 TOPO 2uL | Kan | Lawn of (>200) 0.5mm white colonies |
| S1 TOPO 4 uL (w/ Xgal) | Kan | Lawn of (>200) 0.5mm white colonies |
| S1 P59b (+) ctrl | Kan | ~50 2mm pink colonies, ~50 0.5mm white colonies |
| S1 (-) ctrl | Kan | 50-100 0.5mm white colonies |
It is unclear if the puc origin works because in both sets of transformations, the colonies that grow don't resemble S1 at all. This is at least the 2nd or 3rd instance wherein ColE1 has worked in S1 in P28. The better efficiency of the second set suggests the frozen stocks of S1 competent cells may not be as viable anymore, but we should test this further if we have time before throwing them away-- Shewie take forever to grow to log phase!
Getting Thermoinducible and Lac Inducible GFP into pSC101
Digests of Inducible Systems and pSC101 8/27
P102 needs to be retransformed so it was not included in this set.
Gel of Digest 8/27
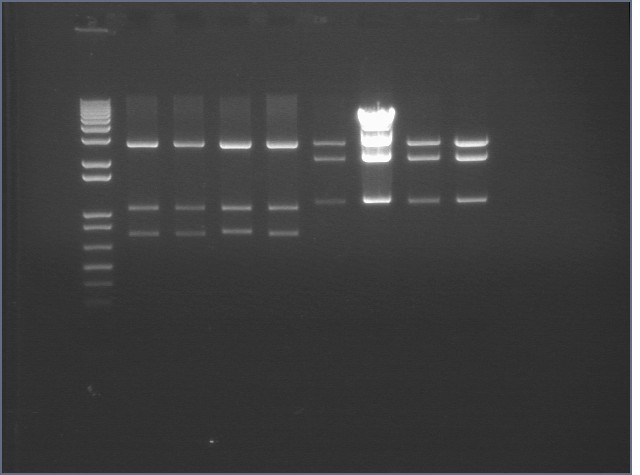
| 1% agarose | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kb + DNA Ladder | |
| 2 | P124 1:2.5 A | |
| 3 | P124 1:2.5 B | |
| 4 | P125 1:2.5 Dephos A | |
| 5 | P125 1:2.5 D | |
| 6 | mtrB TOPO 0.5 uL B | |
| 7 | mtrB TOPO 2 uL B | |
| 8 | mtrB TOPO 4-1 | |
| 9 | mtrB TOPO 4-10 | |
mtrB TOPO bands were cut (2.2 kB) but P124-5 did not cut properly, suggesting that there was an extra cut site that was cutting the insert into two pieces.
Single-cut Digest of P124-5 8/27
IMPORTANT NOTE Further inspection of the sequence revealed that there was an extra PstI site after the cI repressor coding region due to an error that was made while making the primers that resulted in the suffix being backward and the cut sites on the part to read EXPS. Attempts to mutate out the extra site and basepairs will begin next week once primers are reviewed and ordered-- might take a while just to get them.
Due to the extra PstI site, all future ligations involving the thermoinducible system cut with PstI will need to be held off (ie. getting into the pSC101 vector, and adding on mtrB or mtrA) until the issue is resolved either with site directed mutagenesis or by redesigning primers for cI and starting over?
Re-Transformations of Various Plasmids
Scheduled for next week.
Getting mtrB onto thermo and lac inducible systems
Ligating mtrB TOPO with RBS
Grew up more mtrB TOPO for next week. Will need to grow up more P40 and P97 next week then, too.
PCR of mtrB with RBS + mtrB primers
Scheduled for next week.
 "
"