Team:Harvard/Dailybook/Week12/Chemical and Light
From 2008.igem.org
(→Colony PCR 09/27) |
(→Induction test) |
||
| Line 103: | Line 103: | ||
We realized we can't tell whether these are just plain 130 or 38/39+130, so we picked some and grew them in both Amp and Kan. Those that grow only on Amp are in the promoter plasmids. One 39+130 appeared to have worked (no Kan growth, oddly sparse Amp growth). | We realized we can't tell whether these are just plain 130 or 38/39+130, so we picked some and grew them in both Amp and Kan. Those that grow only on Amp are in the promoter plasmids. One 39+130 appeared to have worked (no Kan growth, oddly sparse Amp growth). | ||
==Induction test== | ==Induction test== | ||
| - | + | The 1000x dilutions of samples at 2 hours after induction produced similar results after 5-6 hours. These cells are presumably at a different growth phase from cells that have been growing at much higher concentration for longer, and are thus induced in different conditions. | |
| + | <div style="text-indent:0pt;color:black">[[Image:Lac1000.jpg|720px|thumb|center|Lac Inducibility 1000x Results]] | ||
| + | </div> | ||
Latest revision as of 23:15, 29 October 2008
Contents |
9/22 RE digests
With NEB enzymes, 60 min (buffer 2+BSA):
- 128#1 XPXmnI
- 128#5 XSXmnI
- 128#9 ESXmnI
The XmnI is to cut the backbone, which has a similar length to the mtrB constructs.
With Fermentas enzymes, 25 min:
- 116 SP
- 117 SP
- 108 SP
- 51 EX
- 63 XP
9/23 ligations
The following ligations were performed using 1:6 molar ratio and the Takara kit at 16°C for 45min:
- 128ES+63EX (also did a 5min, 25°C ligation to test whether the extraction method or the ligation conditions are improving the process)
- 128XP+108SP
- 128XP+116SP
- 128XP+117SP
- 51EX+38ES
- Neg ctrl with water as insert
- 117SP+128XS+[63XP]
- 108SP+128XS+[63XP]
- 116SP+128XS+[63XP]
Note that [63] means P63 was added 20 minutes into the ligation (to reduce uptake only of terminator). Prior to addition of ligase, DNA mix was heated to 65°C for 2 minutes and allowed to cool. For ones with 63 added later, the 63 was heated prior to addition, and more ligation mix was added next to make up the volume.
All transformed into DH5α.
9/24 transformation results
Only the P128ES+63EX ligations gave colonies. Both the long and short ligations gave hundreds of colonies, although the long one had more.
9/24 P128+63EX colony PCR
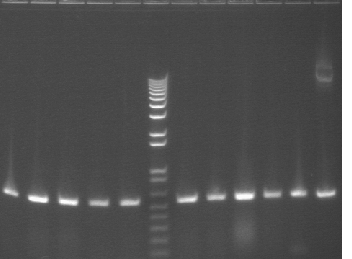
PCR using BBpfx and mtrB-internal500. Should produce bands a little over 500bp. (Numbers on gel correspond to numbers on master plate.)
Everything as expected. New primer works well at 55°C.
Lane 1: Colony 1
Lane 2: Colony 2
Lane 3: Colony 3
Lane 4: Colony 4
Lane 5: Colony 5
Lane 6: 1 kb plus ladder
Lane 7: Colony 6
Lane 8: Colony 7
Lane 9: Colony 8
Lane 10: Colony 9
Lane 11: Colony 10
Lane 12: control
Ligations/Transformations 09/26
25min RE digests (re)done on appropriate minipreps using Fermentas enzymes and extracted using Clonewll gels.
We used the Takara ligation kit to do the following ligations (1:6 vector:insert):
- P38SP + P130XP
- P39SP + P130XP
- P38SP + P51XP
- P116SP + P130XP
- P108SP + P130XP
- P117SP + P130XP
- P51EX + P38ES (1/20)
- P51EX + P38ES (1/100)
- P51EX ligation control
We followed the Takara 25°C protocol and transformed all of the ligations into DH5α. We also let some of the ligations run overnight (16 °C for 8 hours -->12 °C for 4 hours --> 4 °C for eternity).
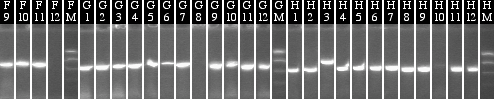
Colony PCR 09/27
Lanes 1-16: P38+51
The band doesn't look bright, but the proper band does appear to be there (should be around 1.4 kb).
Lanes 17-28: P117+130
Lanes 29-48: P116+130
Lanes 49-68: P108+130
These are all wrong- they all look like plain 130 since the band is at 500bp. This means that there must have been uncut 130 in the mix.
Lanes 69-82: P39+130
Lanes 83-96: P38+130
We realized we can't tell whether these are just plain 130 or 38/39+130, so we picked some and grew them in both Amp and Kan. Those that grow only on Amp are in the promoter plasmids. One 39+130 appeared to have worked (no Kan growth, oddly sparse Amp growth).
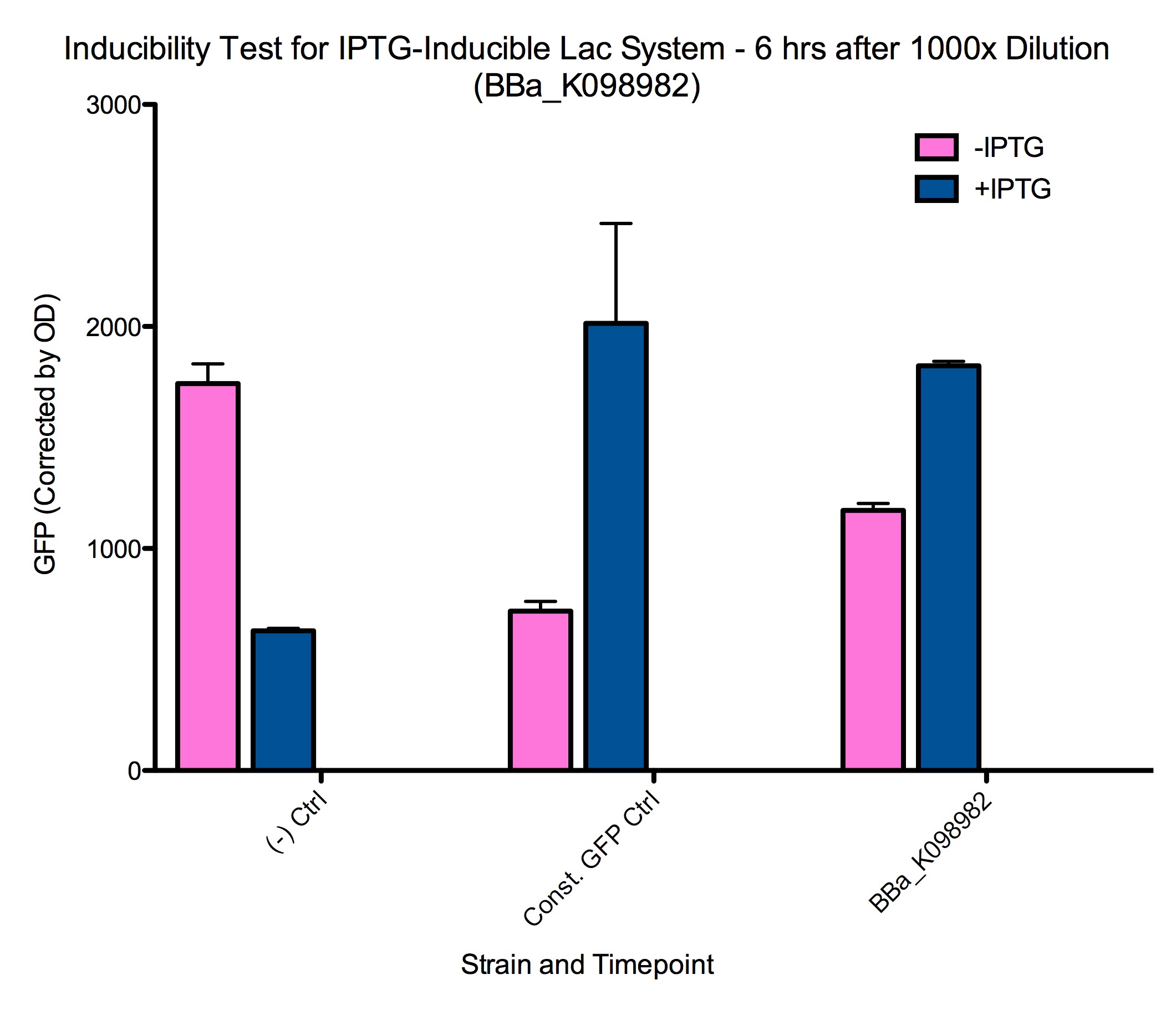
Induction test
The 1000x dilutions of samples at 2 hours after induction produced similar results after 5-6 hours. These cells are presumably at a different growth phase from cells that have been growing at much higher concentration for longer, and are thus induced in different conditions.
 "
"