Team:Hawaii/Project/Part A MaterialsMethodsResults
From 2008.igem.org
m (→PCR amplification of the oriV, rep, and aadA from pRL1383a) |
(→Test of the Plasmid) |
||
| Line 175: | Line 175: | ||
* We need to verify that the plasmid was transferred through conjugation and that it is autonomously replicating. Though ''Synechocystis'' is naturally transformable, plasmids transferred by this pathway cannot re-cyclize, and are only maintained if they integrate into the genome or into one of the endogenous plasmids. A restriction digest of the plasmids isolated from the bacterium can be performed to verify that it is maintained autonomously. | * We need to verify that the plasmid was transferred through conjugation and that it is autonomously replicating. Though ''Synechocystis'' is naturally transformable, plasmids transferred by this pathway cannot re-cyclize, and are only maintained if they integrate into the genome or into one of the endogenous plasmids. A restriction digest of the plasmids isolated from the bacterium can be performed to verify that it is maintained autonomously. | ||
| - | * The copy number of | + | * The copy number of the plasmid with proposed name: pSB7SSt2 will also be determined. |
==References== | ==References== | ||
Latest revision as of 04:01, 30 October 2008
This expression vector will pull parts from the RSF1010 derived broad-host-range plasmid pRL1383a, the self-transmissible plasmid RP4 and also from available BioBrick parts.
The obtained parts will be maintained on seperate plasmids then later compiled onto the BioBrick Base Vector, BBa_I51020. The mobility of this vector will be tested by first transforming into E. coli then through conjugative transferr to Synechocystis PCC6803, followed by the final test of cloning in genes and testing their expression in both E. coli and Synechocystis PCC6803.
Contents |
Step 1: Synthesis and PCR Amplification of Individual Parts
Synthesis and assembly of the oriT, the lac promoter and the RBS
- The origin of conjugative transfer was synthetically constructed using the Overlapping Oligonucleotides method from the Silver Lab [1]. Parts were verified with sequencing.
RP4 Origin of Transfer Sequences for Oligonucleotide Extension
| name | oligonucleotide set | Notes |
|---|---|---|
| oriT1_ob._na.1 | ctagaggaataagggacagtgaagaaggaacacccgctcg | complement oriT4 |
| oriT2_ob._na.1 | cgggtgggcctacttcacctatcctgcccggctgacgccg | complement of oriT5 |
| oriT3_ob._na.1 | ttggatacaccaaggaaagtctacatactagtagcggccgctgca | complement of oriT6 |
| oriT4_ob._na.1 | GCGGCCGCTACTAGTAtgtagactttccttggtg | |
| oriT5_ob._na.1 | tatccaacggcgtcagccgggcaggataggtgaagtaggcc | |
| oriT6_ob._na.1 | cacccgcgagcgggtgttccttcttcactgtcccttattcCT |
Sequencing by the Greenwood Molecular Biology Facility returned the following results:
GAATTTCAGATAAAAAAAATC CTTAGCTTTCGCTAAGGATGA TTTCTGGAATTCGCGGCCGCT TCTAGAGGAATAAGGGACAGT GAAGAAGGAACACCCGCTCGC GGGCAGGCCTACTTCACCTAT CCTGCCCGGCTGACGCCGTTG GATACACCAAGGAAAGTCTAC ATACTAGTAGCGGCCGCTGCA G
Important to note are the correct BioBrick prefix and suffix sequences as well as the correct sequence for the origin of transfer.
Promoter and RBS Sequences for Oligonucleotide Extension
| name | oligonucleotide set | Notes |
|---|---|---|
| B0034-RBS-BkIn_fb._sb.1 | CTAGA G aaagaggagaaa T ACTAGT A GCGGCCG CTGCA | De novo synthesis of BBa_B0034 as back insert, RBS 1.0 |
| B0034-RBS-BkIn_rb._sb.1 | GCGGCCGCTACTAGTAtttctcctctttCT | complement to B0034-RBS-BkIn_fb._sb.1 |
| I14032-PLac-FrIn_fb._sb.1 | AATTC GCGGCCGC T TCTAGA G tggtgcaaaacctttcgcggtatggcatgatagcgcc T A | De novo synthesis of BBa_I14032 as front insert |
| I14032-PLac-FrIn_rb._sb.1 | CTAGTAggcgctatcatgccataccgcgaaaggttttgcaccaCTCTAGAAGCGGCCGCG | complement to I14032-PLac-FrIn_fb._sb.1 |
The Promoter and RBS were ligated and used for further constructions. The sequence and order of the Promoter and RBS were verified later during sequencing of the aadA construct.
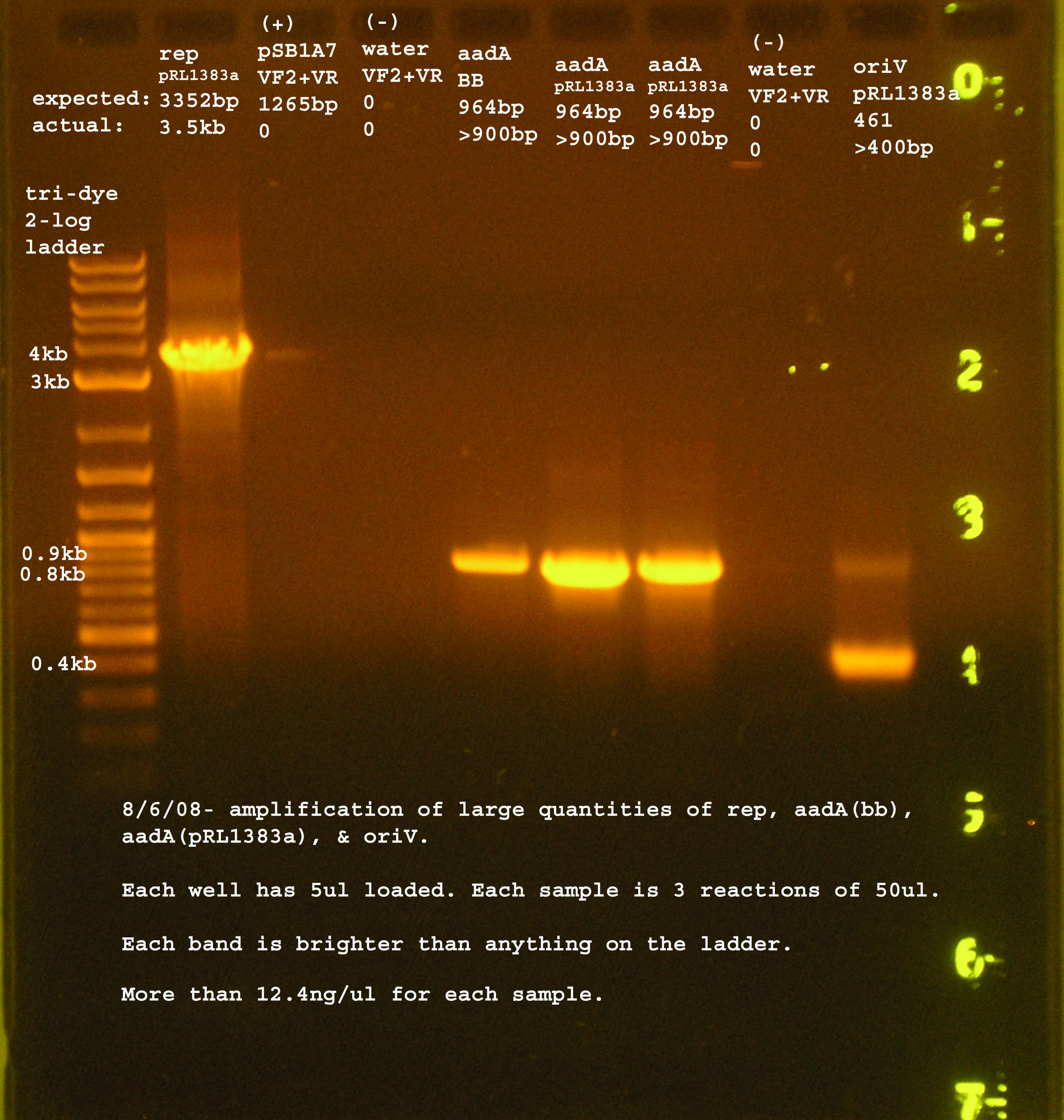
PCR amplification of the oriV, rep, and aadA from pRL1383a
Several regions of pRL1383a will be amplified with BioBrick based primers. These components were used later in the construction of a pRL1383a BioBrick based vector. These parts include the aadA region from pRL1383a and from the BioBrick Registry, the origin of vegetative replication (oriV), and the replication proteins.
pRL1383a Genes w/ BioBrick Ends
| name | primer | Tm | Notes |
|---|---|---|---|
| aadA_fp._sb.1 | cctTTCTAGatgagggaagcggtgatcg | 59.4/65.7 C | isolates aadA from ATG to TAA-TAA |
| aadA_rp._sb.1 | aaggCTGCAGCGGCCGCTACTAGTAttattatttgccgactaccttgg | 55.4/74.5 | isolates aadA from ATG to TAA-TAA |
| pRL1383aOriV_fb._sb.1 | cctTTCTAGAGgaacccctgcaataactgtc | 56.3/65.9 | |
| pRL1383aOriV_rb._sb.1 | aaggCTGCAGCGGCCGCTACTAGTAgctgaatgatcgaccgagac | 58/76.2 | |
| pRL1383aRep_fp._sp.1 | cctTTCTAGatgaagaacgacaggactttgc | 58.9/64.9 | Begins with RepB |
| pRL1383aRep_rb._sb.1 | aaggCTGCAGCGGCCGCTACTAGTAcctatggagctgtgcggca | 62.2/78.5 | Ends RepC terminator.8/2:the terminator is missing the last C. |
Sequencing:
- Each of these parts were ligated into pSB1A3 and transformed into DH5-a cells. The resulting colonies were verified with PCR using VF2 and VR primers. Any positive results were then further verified with sequencing at the Greenwood Molecular Biology Facility.
Step 2: Construction of Plasmid
- The BioBricked parts were then assembled two at a time into pSB1A3. The ligation product was then transformed into DH5-a cells, and the colonies were verified using PCR. Positive PCR results were then sent in to the Greenwood facility for sequencing.
Order of Construction
- Ligation 1:
- The promoter/rbs construct was ligated to aadA--> promoter/rbs/aada
- The promoter/rbs construct was ligated to rep --> promoter/rbs/rep
- Ligation 2:
- promoter/rbs/aada was ligated to oriT --> promoter/rbs/aada/oriT
- promoter/rbs/rep was ligated to oriV --> promoter/rbs/rep/oriV
- Ligation 3:
- promoter/rbs/aada/oriT was ligated to promoter/rbs/rep/oriV --> promoter/rbs/aada/oriT/promoter/rbs/rep/oriV.
- Ligation 4:
- promoter/rbs/aada/oriT/promoter/rbs/rep/oriV + Base Vector --> RSF1010 derived Broad-Host-Range Plasmid
Step 3: Testing of Parts and Plasmid
The construction of the plasmid was not completed, so parts were tested individually. The oriT and aadA are similar to parts already available in the registry, though improvements were made.
Testing of aadA
- AP1 the aadA gene, the the lac promoter and an rbs in pSB1A3. This part is similar to Spectinomycin R open-reading-frame basic part which was built in 2006 and is available in the registry, but the experience states that this part has issues. We checked the sequence of this part and found that it has a point mutation which could be the reason for the lack of expression of this protein.
- The aadA gene submitted by our team has been verified by sequencing and has been shown to grow on LB+Agar plates containing Sm50Sp100.
Test of Conjugation Efficiency of Constructs containing oriT
- The oriTr is the relaxation region for the R plasmid (i.e. RP4) and is available as a part in the registry. We have constructed another version of this relaxation region: oriT.
- We want to compare the efficiencies of these two origins of transfer. To do this, the origins were added to LacZ. After conjugation, the colonies containing the origin of transfer should be identified with blue/white screening and also by antibiotic selection.
- The experiment was originally designed to only use Blue/White screening to assess the efficiency of conjugation for both of the plasmids (oriTr/J33207 and oriT/J33207), however to give more certainty to the results, antibiotic selection was also added.
Test of the Plasmid
- The design of this plasmid allows for testing before it is inserted into the base vector-- all components necessary for conjugation and autonomous replication are present. The construct in pSB1A3 will be transferred through conjugation to Synechocystis PCC6803 using the tri-parental mating method. The final construct will be tested following its completion.
- We need to verify that the plasmid was transferred through conjugation and that it is autonomously replicating. Though Synechocystis is naturally transformable, plasmids transferred by this pathway cannot re-cyclize, and are only maintained if they integrate into the genome or into one of the endogenous plasmids. A restriction digest of the plasmids isolated from the bacterium can be performed to verify that it is maintained autonomously.
- The copy number of the plasmid with proposed name: pSB7SSt2 will also be determined.
References
- Molecular Genetics of Bacteria, 3rd ed., Snyder and Champness (2007) ASM Press
- Scholtz “Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010,” 1988, Gene 75 (1989) 217-288.
- Mermet-Bouvier (1993), “Transfer and replication of RSF1010 derived plasmids in several cyanobacteria in the genera of Synechocystis and Synechococcus,” Current Microbiology 27, 323-327.
- Shetty, Reshma, “Engineering BioBrick vectors from BioBrick parts.” Journal of Biological Engineering 2008, 2:5
- Prentki P, Krisch HM, “In vitro insertional mutagenesis with a selectable DNA fragment,”Gene. 1984 Sep;29(3):303-13.
- Chauvat, “A host-vector system for gene cloning in the cyanobacterium Synechocystis PCC 6803,” Mol. Gen. Genet (1986) 204:185-191.
- Katashkina JI, “Construction of stably maintained non-mobilizable derivatives of RSF1010 lacking all known elements essential for mobilization.” BMC Biotechnol 2007 Nov 21; 7 80.
- Phillips, Ira & Pamela Silver, “A New Biobrick Assembly Strategy Designed for Facile Protein Engineering,” http://hdl.handle.net/1721.1/32535.
- Silver Lab (http://openwetware.org/wiki/Silver:_Oligonucleotide_Inserts)
- Guerry, van Embden, & Falkow 1974
- Bagdasarian et al. 1981
- Wolk et al. “Paired cloning vectors for complementation of mutations in the cyanobacterium Anabaena sp. strain PCC 7120.” Archives of Microbiology 188, 551-563 (2007).
- Vector NTI
 "
"