Team:Heidelberg/Human Practice/Phips the Phage/Project Details
From 2008.igem.org
Annastoeckl (Talk | contribs) (→Project Details) |
Annastoeckl (Talk | contribs) (→Sensing: Cell Recognition and Targeting) |
||
| (9 intermediate revisions not shown) | |||
| Line 482: | Line 482: | ||
== '''Sensing: Cell Recognition and Targeting''' == | == '''Sensing: Cell Recognition and Targeting''' == | ||
| - | In the following I will first explain you the detection system of the killer strain and after that the killing system. | + | In the following I will first explain to you the detection system of the killer strain and after that the killing system. |
| - | At first my friends had to design the prey strain as the artificial pathogen (working with real pathogens in the lab would have been | + | At first my friends had to design the prey strain as the artificial pathogen (working with real pathogens in the lab would have been too dangerous for my student friends). For that they inserted (we say transformed) a gene into this strain which is responsible for the secretion of a specific pathogen-derived molecule. The team chose the gene LuxS from the bacterium ''Vibrio herveji'' encoding for a protein (an enzyme) able to produce a molecule called autoinducer II. This autoinducer symbolized the molecules a pathogen secretes and was in the following used as the signal molecule the killer strain should be able to detect. Engineering the prey strain was very simple and autoinducer production was easy to achieve. <br> |
| + | [[Image:Sensing_phips1.png|thumb|900px|The chemotaxis receptor recognizes nutrient molecules like amino acids and sends a signal to the motility apparatus of the bacterium. The quorum sensing receptor recognizes autoinducer molecules.]] | ||
| + | <br> | ||
The more difficult part of the project was the detection of these signal autoinducers by the killer strain, so that the killer would recognize the artificial pathogen strain and swim in direction of it. | The more difficult part of the project was the detection of these signal autoinducers by the killer strain, so that the killer would recognize the artificial pathogen strain and swim in direction of it. | ||
| - | To achieve this purpose, the team engineered a strain containing an artificial chemotaxis receptor | + | To achieve this purpose, the team engineered a strain containing an artificial [[Team:Heidelberg/Human_Practice/Phips_the_Phage/General_Backround#Chemotaxis|chemotaxis]] receptor. The important step of this system was the detection of signal molecules (in nature this would be for example nutrient molecules) so that the bacterium would swim in direction of highest concentration of these molecules- and by that to find the source where all nutrients come from. |
| - | The detection of the nutrient molecules | + | The detection of the nutrient molecules are mediated by the chemotaxis receptor called tar. This receptor consists mainly of two different components: The part responsible for recognizing the nutrient molecule and the signal sending part, which is responsible for activating bacterial swimming in direction of the nutrient source. |
My scientist friends now wondered, whether it would be possible to exchange the component of the receptor responsible for recognizing the nutrient molecule (the ligand binding domain) by something able to recognize the autoinducer II- the signal molecule the artificial pathogens secreted. <br> | My scientist friends now wondered, whether it would be possible to exchange the component of the receptor responsible for recognizing the nutrient molecule (the ligand binding domain) by something able to recognize the autoinducer II- the signal molecule the artificial pathogens secreted. <br> | ||
| - | And they found something in nature- another receptor- able to recognize just that autoinducer molecule. In addition this receptor had nearly the same structure as the chemotaxis receptor: It also consisted of a ligand binding | + | And they found something in nature- another receptor- able to recognize just that autoinducer molecule. In addition, this receptor had nearly the same structure as the chemotaxis receptor: It also consisted of a ligand binding part and a function-triggering part. This receptor is the Quorum-Sensing receptor from ''Vibrio harveji''. <br> |
<br> | <br> | ||
| - | [[Image: | + | [[Image:Sensing_phips2.png|thumb|600px|Cloning strategy to make a fusion receptor. First the needed parts of the receptor genes (the recognition part of the quorum sensing receptor gene and the signal sending part of the chemotaxis receptor gene) have to be isolated. Then they are put together and brought into a plasmid. This is transformed into bacteria cells where the fusion protein is produced. Thereby the bacteria are enabled to swim into the direction of autoinducers, using the chemotaxis motility system.]] |
| - | + | ||
| - | When the team recognized the similarity | + | When the team recognized the similarity between the chomotaxis receptor tar and the quorum sensing receptor LuxQ they thought it should be possible to exchange the ligand binding domain of tar (the chemotaxis receptor) by the ligand binding domain of LuxQ (the quorum sensing receptor). And that is what they did. <br> |
| - | Via PCR the Team amplified the ligand binding domain of LuxQ and the swimming-triggering (signal transferring) domain of tar in a first reaction. In a second PCR the team fused the two gene fragments of LuxQ and tar and thus got a LuxQ-Tar fusion receptor- a so called chimeric receptor. The exact PCR strategy is quite complicate- but the figure below explains the strategy in a simplified | + | Via PCR the Team amplified the ligand binding domain of LuxQ and the swimming-triggering (signal transferring) domain of tar in a first reaction. In a second PCR the team fused the two gene fragments of LuxQ and tar and thus got a LuxQ-Tar fusion receptor- a so called chimeric receptor. The exact PCR strategy is quite complicate- but the figure below explains the strategy in a simplified form. <br> <br> |
| - | Perhaps you know chimera from myths and fairy stories your grand parents told you. Chimera are fabulous creatures, often mixtures between humans and animals, i.e. a horse with a human | + | Perhaps you know chimera from myths and fairy stories your grand parents told you. Chimera are fabulous creatures, often mixtures between humans and animals, i.e. a horse with a human head (Harry Potter loves them!). |
| - | You can imagine my friends’ receptor being built the same way: | + | You can imagine my friends’ receptor being built the same way: Head and arms from LuxQ (to catch the signalling molecules) and body with feet from the chemotaxis receptor (to move in direction of the artificial pathogen). So the direction, the bacteria containing the chimeric receptor will choose to swim in, is no longer controled by the original chemotaxis receptor, but by the way the LuxQ-part of the chimeric receptor ordains to our bacteria. <br> |
<br> | <br> | ||
| - | + | ||
| - | + | Instead of following nutrient molecules in the environment the killer bacteria would now follow the autoinducer molecules. And by following the autoinducer gradient our killer strain would finally reach the artificial pathogens. | |
| - | Instead of following nutrient molecules in the environment the killer bacteria would now follow the autoinducer molecules. And by following the autoinducer gradient our killer strain would | + | Quite astonishing, right? Now that you gained an insight into the sensing module I will show you another exciting part of the project: the killing. |
| - | Quite astonishing, right? | + | |
<br> | <br> | ||
[[https://2008.igem.org/Team:Heidelberg/Human_Practice/Phips_the_Phage/Project_Details back]] | [[https://2008.igem.org/Team:Heidelberg/Human_Practice/Phips_the_Phage/Project_Details back]] | ||
| - | + | <br> | |
| + | <br> | ||
== '''Phage project''' == | == '''Phage project''' == | ||
| Line 580: | Line 581: | ||
== '''Colicin Project''' == | == '''Colicin Project''' == | ||
| - | In many habitats of bacteria nutrients and other substances which bacteria need for living are quite rare. For this reason the different bacteria strains living there compete and fight each other to ensure the surviving of the own strain. These habitats are like some really bad districts in Chicago in the 1920s and 1930s, where different mafia families fought for the dominance in the districts. Like the mafia in these years, there are also some really bad guys among the bacteria, which do not hesitate to kill their competitors if this brings an advantage to them or to their strain. Therefore some bacteria developed the ability to produce toxins, so called bacteriocins. For ''E. coli'' these toxic proteins are called “colicins”, because they are bacterio'''cins''' and produced by ''E. '''''coli''' bacteria. There are many different ways how colicins act, but the result is always the same: They kill any foreign competitors which do not produce the immunity protein to this specific colicin. This immunity protein is like a bulletproof west and is only produced by the bacteria strain also producing the corresponding colicin. It interacts with the colicin and ensures by this, that the other bacteria of this strain are not killed if one bacterium starts the colicin production. In contrast to the immunity proteins, the colicins are not produced all the time. Their production is regulated by a so called SOS promoter. The activation of this promoter can be seen like a provokation of a one mafia family to another or a fight of food, when there is not enough to eat. So the promoter only starts colicin production if the ''E. colis'' get stressed. This can happen for example if there are not enough nutrients or if there are too many other bacteria around. So better never stress an ''E. coli''! And I am really serious about this, because if you think “what can such a small organisms harm me?” you are really on the wrong track. Researches could show that some kinds of colicin proteins are not only able to harm prokaryotic cells like bacteria but also eukaryotic cells like human cells! Thus researches attempt to make use of this dangerous discovery by trying to fight dangerous body-own eukaryotic cells | + | In many habitats of bacteria nutrients and other substances which bacteria need for living are quite rare. For this reason the different bacteria strains living there compete and fight each other to ensure the surviving of the own strain. These habitats are like some really bad districts in Chicago in the 1920s and 1930s, where different mafia families fought for the dominance in the districts. Like the mafia in these years, there are also some really bad guys among the bacteria, which do not hesitate to kill their competitors if this brings an advantage to them or to their strain. Therefore some bacteria developed the ability to produce toxins, so called bacteriocins. For ''E. coli'' these toxic proteins are called “colicins”, because they are bacterio'''cins''' and produced by ''E. '''''coli''' bacteria. There are many different ways how colicins act, but the result is always the same: They kill any foreign competitors which do not produce the immunity protein to this specific colicin. This immunity protein is like a bulletproof west and is only produced by the bacteria strain also producing the corresponding colicin. It interacts with the colicin and ensures by this, that the other bacteria of this strain are not killed if one bacterium starts the colicin production. In contrast to the immunity proteins, the colicins are not produced all the time. Their production is regulated by a so called SOS promoter. The activation of this promoter can be seen like a provokation of a one mafia family to another or a fight of food, when there is not enough to eat. So the promoter only starts colicin production if the ''E. colis'' get stressed. This can happen for example if there are not enough nutrients or if there are too many other bacteria around. So better never stress an ''E. coli''! And I am really serious about this, because if you think “what can such a small organisms harm me?” you are really on the wrong track. Researches could show that some kinds of colicin proteins are not only able to harm prokaryotic cells like bacteria but also eukaryotic cells like human cells! Thus researches attempt to make use of this dangerous discovery by trying to fight dangerous body-own eukaryotic cells, for example cancer cells with colicins. These approaches are only at the beginning, but hopefully we will have a therapy against an infection or against cancer based on colicins in the future. |
[[https://2008.igem.org/Team:Heidelberg/Human_Practice/Phips_the_Phage/Project_Details back]] | [[https://2008.igem.org/Team:Heidelberg/Human_Practice/Phips_the_Phage/Project_Details back]] | ||
Latest revision as of 16:59, 29 October 2008


Project Details
Hi, I’m very proud of you- today you really took the challenge to learn something interesting and - I have to confess that - not easy at all. But in the following I’ll try to introduce you into the details of the Heidelberg iGEM project- and I bet that it will be lots of fun.
Until now you got a large background knowledge on genetic engineering, synthetic biology and laboratory techniques.
You also heard that the team of my scientist friends tries to engineer killer bacteria to terminate pathogens or even cancer cells. I will tell you more about killing cancer cells in a separate section just at the bottom of this page under “future perspectives”. Cancer cells and tumors in general are so special, that they need a separate introduction- and I think it’s worth it!
But although my friends made good progress in the project, they were of course not able to develop a perfect medical application in only three months. The question comes up: What should they do as a project if the time is too short to develop a real medical application? Well, my friends thought that the interesting thing about their project is the concept behind it- not only the application.
Thus they wanted to proof that their concept is practicable and well thought through. So they created a model system in the lab to build up a “proof-of-principle” system.
For that they engineered two different bacteria strains: One strain behaving like pathogenic bacteria, which are in nature able to cause harm to human beings. They called this strain the prey strain, because this strain should later on be detected and killed.
They engineered a second bacterial strain, containing a detection system able to locate the prey strain. My scientist friends called this strain the killer strain, because it should later be able to kill the prey strain effectively after a successful detection process.
The teams’ project was divided into two modules: The construction of the prey strain to make it behaving like a pathogen bacteria and the construction of the killer strain. The construction of the killer itself was again divided into sub modules: The creation of the detection system and the development of the killing system.
At least, the iGEM team Heidelberg hoped to develop a system, where the killer strain would be able to detect the prey strain, swim in direction of the prey cells and then kill them specifically. To simplify the model system you could call it predator-prey model.
Sensing: Cell Recognition and Targeting
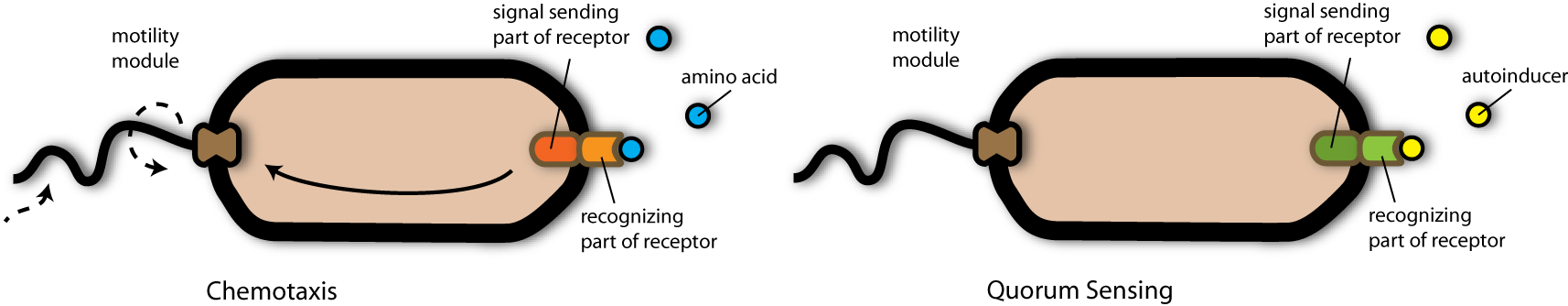
In the following I will first explain to you the detection system of the killer strain and after that the killing system.
At first my friends had to design the prey strain as the artificial pathogen (working with real pathogens in the lab would have been too dangerous for my student friends). For that they inserted (we say transformed) a gene into this strain which is responsible for the secretion of a specific pathogen-derived molecule. The team chose the gene LuxS from the bacterium Vibrio herveji encoding for a protein (an enzyme) able to produce a molecule called autoinducer II. This autoinducer symbolized the molecules a pathogen secretes and was in the following used as the signal molecule the killer strain should be able to detect. Engineering the prey strain was very simple and autoinducer production was easy to achieve.
The more difficult part of the project was the detection of these signal autoinducers by the killer strain, so that the killer would recognize the artificial pathogen strain and swim in direction of it.
To achieve this purpose, the team engineered a strain containing an artificial chemotaxis receptor. The important step of this system was the detection of signal molecules (in nature this would be for example nutrient molecules) so that the bacterium would swim in direction of highest concentration of these molecules- and by that to find the source where all nutrients come from.
The detection of the nutrient molecules are mediated by the chemotaxis receptor called tar. This receptor consists mainly of two different components: The part responsible for recognizing the nutrient molecule and the signal sending part, which is responsible for activating bacterial swimming in direction of the nutrient source.
My scientist friends now wondered, whether it would be possible to exchange the component of the receptor responsible for recognizing the nutrient molecule (the ligand binding domain) by something able to recognize the autoinducer II- the signal molecule the artificial pathogens secreted.
And they found something in nature- another receptor- able to recognize just that autoinducer molecule. In addition, this receptor had nearly the same structure as the chemotaxis receptor: It also consisted of a ligand binding part and a function-triggering part. This receptor is the Quorum-Sensing receptor from Vibrio harveji.
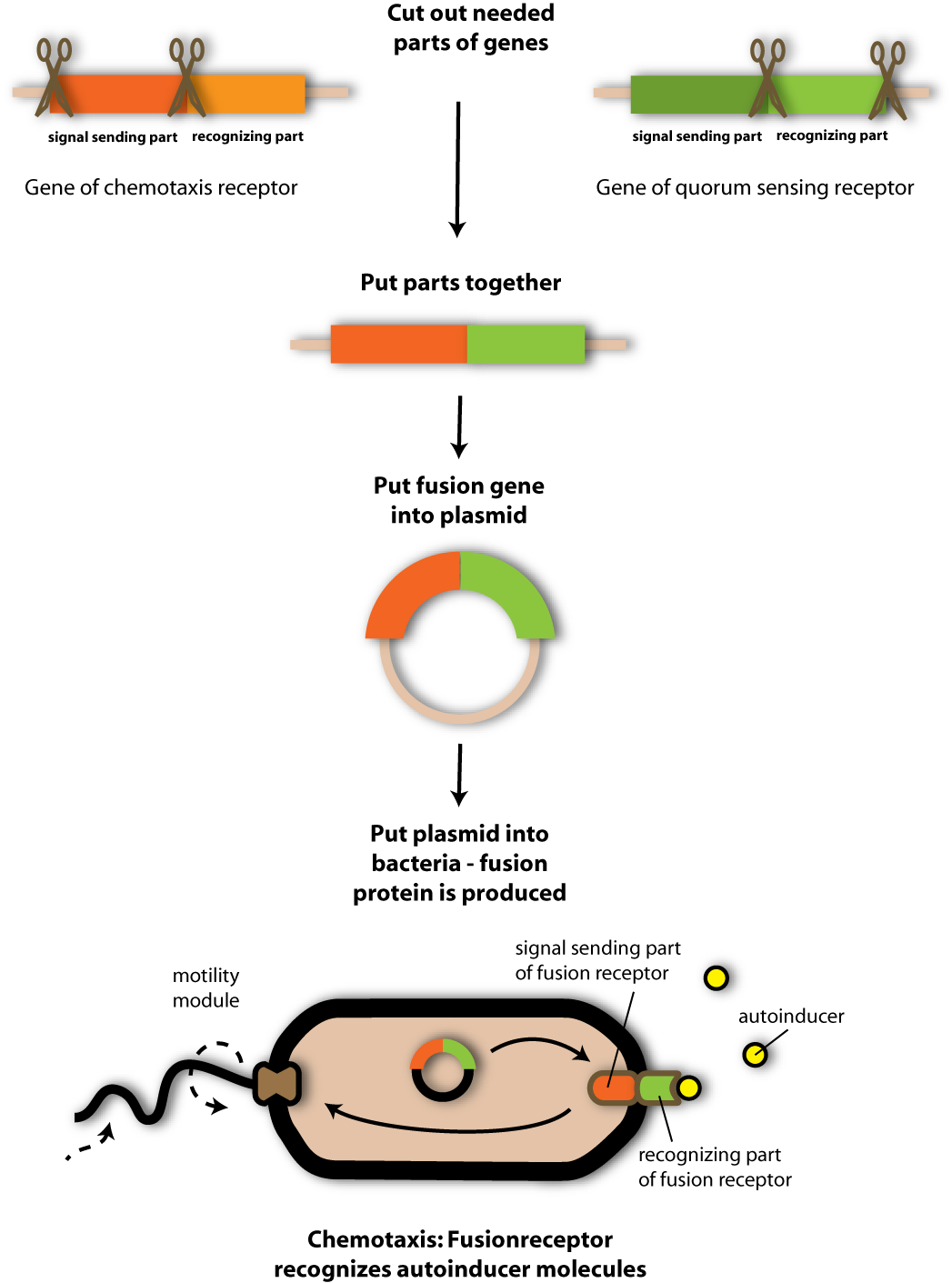
When the team recognized the similarity between the chomotaxis receptor tar and the quorum sensing receptor LuxQ they thought it should be possible to exchange the ligand binding domain of tar (the chemotaxis receptor) by the ligand binding domain of LuxQ (the quorum sensing receptor). And that is what they did.
Via PCR the Team amplified the ligand binding domain of LuxQ and the swimming-triggering (signal transferring) domain of tar in a first reaction. In a second PCR the team fused the two gene fragments of LuxQ and tar and thus got a LuxQ-Tar fusion receptor- a so called chimeric receptor. The exact PCR strategy is quite complicate- but the figure below explains the strategy in a simplified form.
Perhaps you know chimera from myths and fairy stories your grand parents told you. Chimera are fabulous creatures, often mixtures between humans and animals, i.e. a horse with a human head (Harry Potter loves them!).
You can imagine my friends’ receptor being built the same way: Head and arms from LuxQ (to catch the signalling molecules) and body with feet from the chemotaxis receptor (to move in direction of the artificial pathogen). So the direction, the bacteria containing the chimeric receptor will choose to swim in, is no longer controled by the original chemotaxis receptor, but by the way the LuxQ-part of the chimeric receptor ordains to our bacteria.
Instead of following nutrient molecules in the environment the killer bacteria would now follow the autoinducer molecules. And by following the autoinducer gradient our killer strain would finally reach the artificial pathogens.
Quite astonishing, right? Now that you gained an insight into the sensing module I will show you another exciting part of the project: the killing.
[back]
Phage project
The phage project is concerned with engineering a module, which is able to kill pathogenic bacteria. These can be species, which invade wounds, for example, and lead to an infection, which can, in the worst case, end with the death of the patient due to a septic inflammation. There are also other bacterial infections inside the human body like pneumonia, for which our method could be a possible treatment.
Since bacteriophages are very efficient natural devices to destroy bacterial populations (as described in the virus section), we decided to use the power nature already invented for their project. But why is it reasonable to invent something new to kill bacteria, since we already have very powerful tools to do this: antibiotics? Well, you may have heard on the news, that there are many pathogenic bacterial strains that have become resistant to antibiotics, like the one that causes pneumonia. The diseases they cause cannot be cured with antibiotics any more.
The general idea is to bring a bacteriophage into a pathogenic bacterial cell. There it will be able to carry out its intrinsic killing mechanism to destroy this cell. A very important thing to our team was that the phages are specifically transferred to pathogen cell – because otherwise one could just take bacteriophages and apply them on the site of bacterial infection. This feature of targeting will be carried out by the bacteria the ‘sensing group’ of our team engineered.
The tasks the phage group had to carry out were to engineer the bacteriophages in a way that it could be stably packed into the ‘killer bacterium’ (the one that will kill the pathogen) without killing this useful cell and find a method to transfer the phage into the pathogenic cell, where it should get active and kill the pathogen.
We will go through these steps in a very general way to see, how the engineering work has been done and how this system works. For detailed information see the project description of this group.
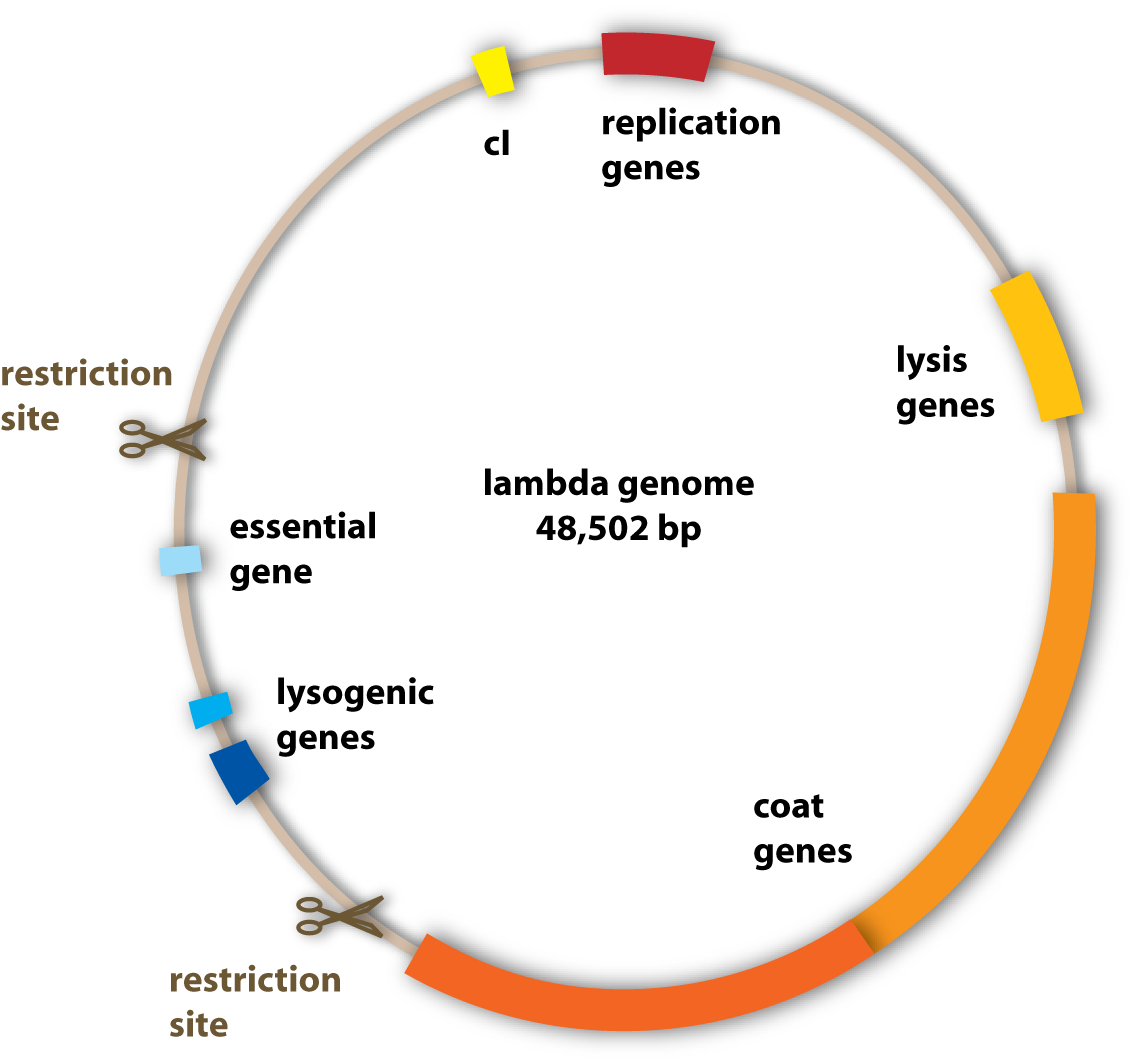
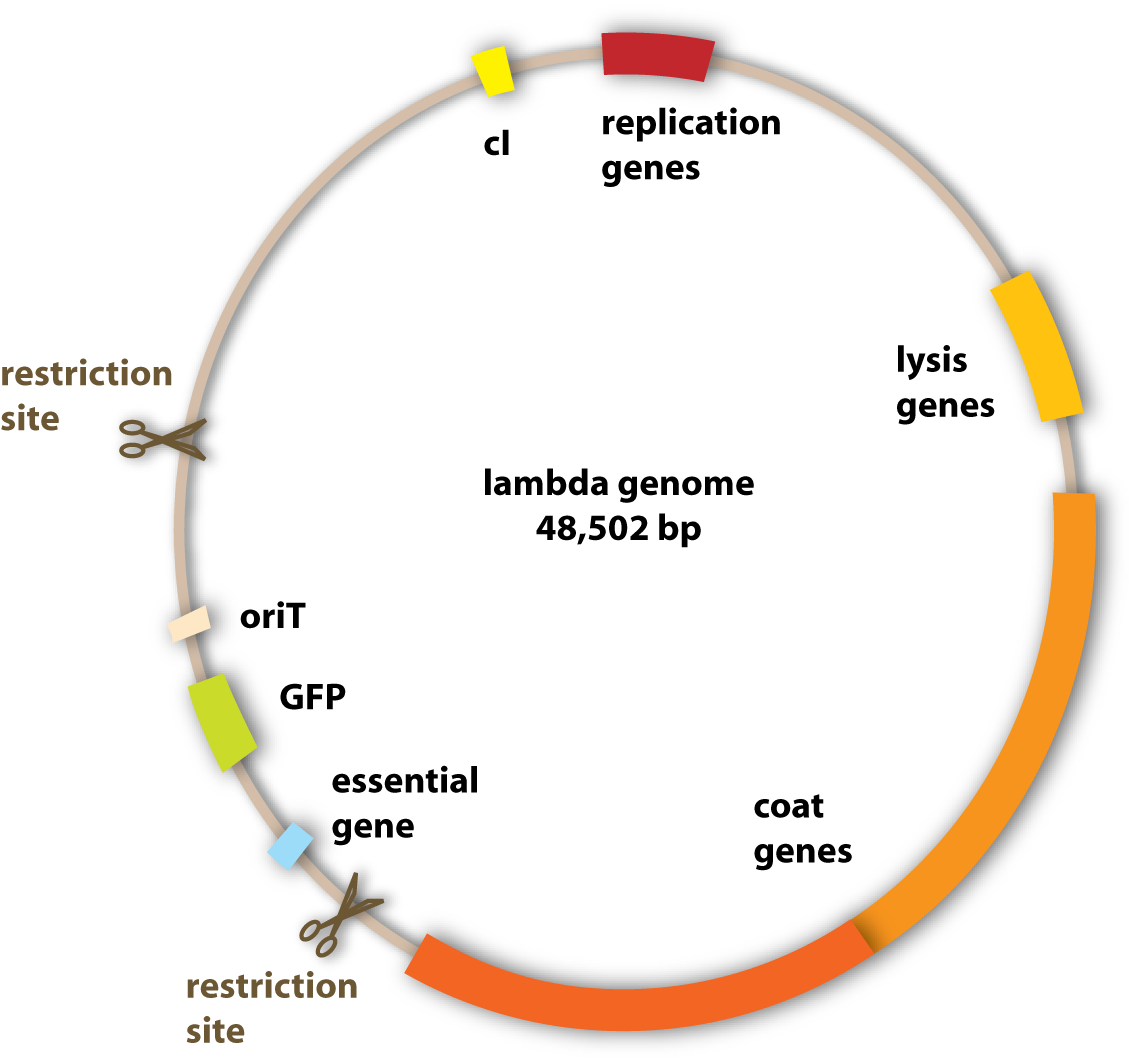
To change the properties of the lambda phage – to make it stay stably in the ‘killer’ cells for example, the genome of the phage has to be changed. This is because the genome codes for all the proteins of the phage and these are responsible for the shape, the replication and the lysis of the host cells. To change the properties of these proteins, the underlying genes have to be changed. This can be done by genetic engineering using standard molecular cloning techniques.
What we wanted to get rid of specifically were the genes which encoded for the lysogenic property of the phage (see section viruses). Since we want to engineer a very efficient killing device, it would not be very reasonable to have a bacteriophage, which is able to integrate into the pathogen’s genome and stay there without killing it.
The properties we wanted to add to the phage were one to be able to transfer it to the pathogen and one to make it fluorescent to visualize it.
Generally, the genome of the lambda phage can be handled like a standard plasmid vector. But working with the genome of the lambda phage is far from being trivial, even to experienced scientists. The difference to the common lab work is that the lambda genome is huge compared to ordinary plasmids, genetic engineers work with: It is 10 times as big as many standard plasmids. And there is another big problem: Since the lambda genome is not an optimized tool for genetic lab work, it contains few useable restriction sites. Remember, these are the sites, where you can cut a DNA-strand – to put some new base pairs (and thereby information) into the sequence or to cut some out. But you can only modify such a huge DNA in the given time we had by molecular cloning. Therefore we had to cut out much bigger fragments than we wanted to, which still contained genes we needed, because we wanted to discard some genes from the genome, and therefore we had to use the few restriction sites available, even if they didn’t fit our purposes very well. The genes we unwontedly cut out had to be put into the phage genome again together with the other genes we wanted to add, to give the phage new properties. [back]
How can the lambda phage bee detained of lysing the killer bacteria?
There is a protein called the lambda repressor or cI, which completely represses the activity of the lambda phage in a bacterial cell. This protein is naturally made by the lambda phage under certain conditions. It binds to the genome of the phage and prevents the transcription of the genes necessary for the induction of an infection cycle: It inhibits the RNA-polymerase from binding to the lambda DNA. No proteins can be made, which means that the phage cannot carry out any activity (see section Genes and Proteins). But why does the lambda phage make such a protein naturally, since it can repress its own activity?
Well, the lambda phage uses it to repress other lambda phages. If one phage has infected a bacterial host and carries out a lysogenic cycle, it produces this protein. Since it is not active in this mode anyway, it does not matter for itself. But it is important, if other bacteriophages infect this particular host cell. Their activity is repressed from the beginning on and they neither can carry out lytic nor lysogenic cycle. Therefore their DNA will just be degraded by the host cell. So the original invader defends and protects its host cell against other lambda phages with the help of the lambda repressor protein.
If we now bring this protein into our killer cells, the lambda phage we will later put in will be inactive and can not kill our engineered cells. [back]
How can we make the phage fluorescent?
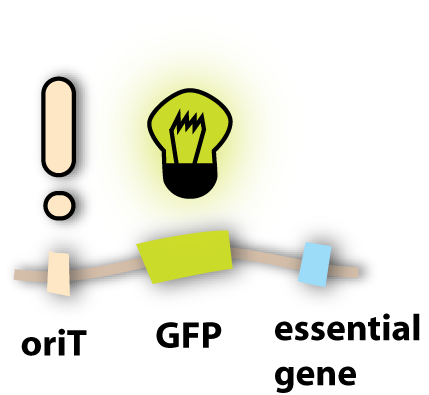
Well, to be precise, we do not make the phage fluorescent, but the phage will give this property to all cells where it is in. This property is gained by using fluorescent proteins. They are able to absorb light and emit light of lower energy and thereby glow. One protein with this feature is taken from a jellyfish and called Green Fluorescent Protein (GFP). It is the oldest and most broadly used fluorescent protein in biotechnology. Because it had such a great impact on this science and improved it a lot, this year’s Nobel Prize in chemistry was awarded for the discovery of GFP and its applications. If GFP is made by a cell and you illuminate this cell with UV-light, you can see a green light coming from this cell. In adding the gene for GFP to the genome of the lambda phage, the cell hosting this phage is able to produce GFP and can thereby be detected. [back]
How can we make the phage transferable to the pathogen?
Generally, we do not want to transfer the phage in its extracellular form to the pathogen, in which the DNA is enclosed in the protein coat and the bacteriophage is able to infect the pathogen in its own. Because to set this form free, the host cell of the phage has to be lysed. We do not intend to destroy our killer cells to set the phage free, because then they could only be active for a short period of time. In finding a mechanism to transfer the phage without the lysis of the host cell, the killer bacteria could be active over a longer period of time and therefore really kill all the pathogenic bacteria
So the idea we had was not to transfer the whole phage, but only its genome, since this has all the properties needed to lyse the target cell. A very powerful natural tool to transfer genetic information is bacterial conjugation (link). So the thing to do was to make the lambda genome accessible for conjugation. Normally, as I explained in the conjugation section, there are conjugative plasmids, which carry all the genes needed for their own conjugation. Since these are quite a lot of genes, we did not want to put them all into the lambda genome. But we found an easier solution: Binary vector systems for conjugation.
Most of the proteins, which are essential for conjugation, do not have to be on the plasmid that is transferred. This makes sense if one imagines that the proteins are produced in the bacterial cell and can spread there and get to the working places. So it doesn’t matter if a conjugative protein is produced from the chromosome, from an additional plasmid or from the plasmid which will be transferred. So what we had to do was to take a conjugative plasmid, which produces all the essential conjugative genes and put it in the killer cells?
But how do these conjugative proteins know which plasmid they should transfer? For this task, there exists a sequence in the plasmid to be transferred, which is called oriT. It tells the conjugative proteins: ‘Hey guys, this is the plasmid you are looking for!’ and provides a site for them to bind to and to get active. So in adding this oriT site to the lambda genome, the phage DNA can be transferred to other bacterial cells. [back]
Summary
And remember the reason why it is stable in the host cell and not lysing it? Because of the expression of the lambda repressor protein! But this protein is not expressed in the target pathogen cell. So once transferred there, the phage DNA can be transcribed, and the proteins for the lytic cycle can be made. There is no possibility to carry out the lysogenic cycle, because we removed the genes necessary for this mode from the genome. So the lambda phage will carry out its work in the pathogen and destroy it.
And now comes the best part of this system: As I already told you, phages are multiplying during the lytic cycle. So when the pathogen cell is destroyed, approximately one hundred new phages are release (originating from one infecting phage). So the number of phages is multiplying with every round of infection: We are having a Domino Effect. This clearly contributes to the aim to kill the whole population of pathogenic bacteria in the inflammatory tissue.
[back]
Colicin Project
In many habitats of bacteria nutrients and other substances which bacteria need for living are quite rare. For this reason the different bacteria strains living there compete and fight each other to ensure the surviving of the own strain. These habitats are like some really bad districts in Chicago in the 1920s and 1930s, where different mafia families fought for the dominance in the districts. Like the mafia in these years, there are also some really bad guys among the bacteria, which do not hesitate to kill their competitors if this brings an advantage to them or to their strain. Therefore some bacteria developed the ability to produce toxins, so called bacteriocins. For E. coli these toxic proteins are called “colicins”, because they are bacteriocins and produced by E. coli bacteria. There are many different ways how colicins act, but the result is always the same: They kill any foreign competitors which do not produce the immunity protein to this specific colicin. This immunity protein is like a bulletproof west and is only produced by the bacteria strain also producing the corresponding colicin. It interacts with the colicin and ensures by this, that the other bacteria of this strain are not killed if one bacterium starts the colicin production. In contrast to the immunity proteins, the colicins are not produced all the time. Their production is regulated by a so called SOS promoter. The activation of this promoter can be seen like a provokation of a one mafia family to another or a fight of food, when there is not enough to eat. So the promoter only starts colicin production if the E. colis get stressed. This can happen for example if there are not enough nutrients or if there are too many other bacteria around. So better never stress an E. coli! And I am really serious about this, because if you think “what can such a small organisms harm me?” you are really on the wrong track. Researches could show that some kinds of colicin proteins are not only able to harm prokaryotic cells like bacteria but also eukaryotic cells like human cells! Thus researches attempt to make use of this dangerous discovery by trying to fight dangerous body-own eukaryotic cells, for example cancer cells with colicins. These approaches are only at the beginning, but hopefully we will have a therapy against an infection or against cancer based on colicins in the future. [back]
Cancer
As you might perhaps have already seen, the iGEM Team Heidelberg project deals with killing nasty cells: pathogen bacteria or even cancer cells. You have probably heard of the cancer disease as it is one of the most dangerous and harmful diseases existing on earth. More than 13 % of the people in the world die from the consequences of cancer. But that should not frighten you too much. Today there are already many very successful-working therapies for this disease, as chemotherapy, radiation therapy and hormone therapy. A problem common for all these therapies is, that they normally affront systematically, which means that every cell of the human body, if it was a cancer cell or not would be effected by the therapy method.
There are several modern approaches to construct pharmaceuticals which take advantage of specific features of cancer cells- but none of them is specific enough to prevent strong side effects in therapy. And that is, where our iGEM project comes into game. But before I tell you more about the idea the Heidelberg project is based on, I will give you some background on cancer development. The most important question is of course:
What is cancer?
Well, cancer is the topic title of many diseases with quite different symptoms. You can find cancer in the blood, liver, loung- well, just everywhere you can imagine. Some forms are good (we say benign), which means that they stay where they were born, others are bad, so called malign cancer and spread out through the whole body. Many sorts of cancer lead to the formation of solid tumors, but there are also forms, like Leukaemia (blood cancer), which are just widespread, single tumor-cells unable to form dense and solid structures. So the first thing you learned about cancer is, that there are different forms of it, which can be found everywhere in the body system.
But the answer remains: what is cancer? To answer this question we can at fist have a view on the opposite question: What is cancer not? Cancer is not a bacterium, virus or other biological, DNA-containing particle. Cancer is pandered by outer influences but the disease itself is home-made, which means that the human body itself forms cancer.
So the first question arising is: what is cancer? Cancer is the accumulation of non-natural, genetically mutated cells, which are optimized for only one single function: Growth. And although you might at first wonder a bit: “Hey, I am also growing every day and I feel fine up to date” we assure you: Growth is never good, except when being under strict regulation and high level of control by cellular control mechanisms. And this information already guides you to the reason why a normal body-cell might derive into a cancer cell: The mechanisms controlling cellular growth are destroyed or full time off-state.
So how does the normal cellular growth control function?
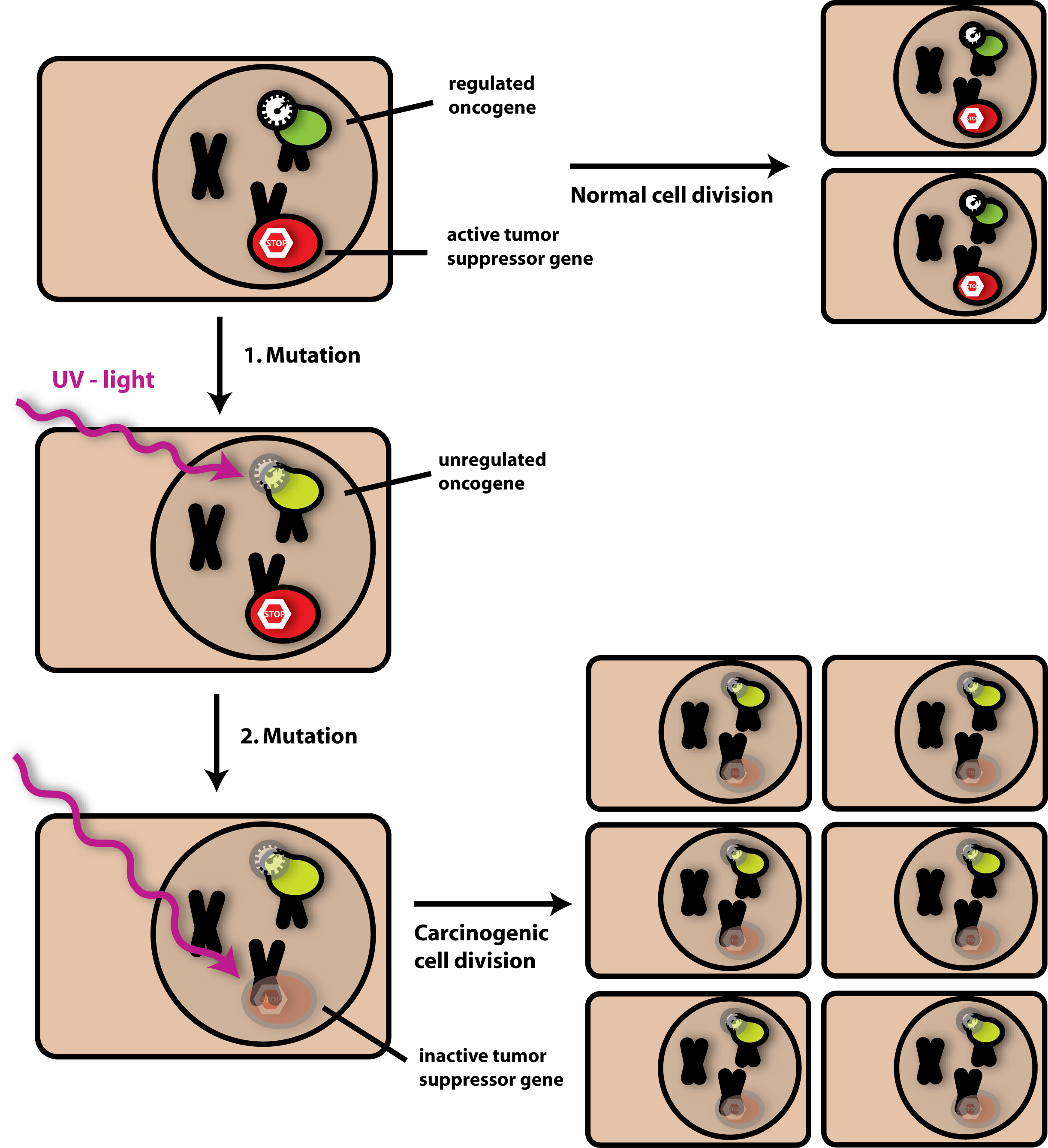
As you already learned all cells existing on earth contain DNA, storing all information necessary to built up an organism. In human cells there are chromosomes containing 20.000 genes. And every gene has a certain function. In context of growth control there are mainly two different genes of certain interest: So called oncogenes and tumorsupressor genes. Oncogenes are genes coding for growth factors. When active, oncogenes lead to a continous cell growth and by that to cellular reproduction. Tumorsupressor genes are genes with a regulatory function. That means when activated they stop cell growth, even if there were growth factors present in the cell. Thus, the cell has a double-secure mechanism which controls cellular reproduction. Of course there are many more of these regulatory circles in the cell, but for a basic understanding we can assume cell growth to be controlled by two genes.
If you remember what I said at the beginning about cancer and cell growth you already know what is coming next. To enable cancer development the tumorsupressor gene and the oncogene have to be manipulated. At this point there is the outer influence coming into game. A problem about our genome is that the information is stored in a material, which is relatively stable, but which can be highly damaged under certain conditions. This damage is called mutation, because it changes the DNA base sequence and by that possibly the information encoded on the DNA strand. Sun Light, so called ultraviolet light for example is able to destroy DNA by building base-dimers. That means that neighboured bases get linked to each other in a strong way which prevents the cell from reading out the information correctly. Other factors like radiation, hormone imbalance or chemicals, and – you should remind that- cigarette smoke are agents, which can have similar effects. Ethidium bromide, which you already got to know in the section Gelelectorphoresis in the Technical Backgound part is such a chemical agent called intercalator. Intercalators are substances intergrating into the DNA double strand and damaging it that way. In case of Ethidium bromide we took advantage of this property by using it as a staining substance for visualizing DNA samples on the gel.
And now, imagine that your DNA is damaged just in the region of a tumor suppressor gene. The consequence would at first not be very dramatic, because your oncogenes would still be well controlled. But if a second mutation step would mutate your oncogene, too, and if this mutation caused continuous, rapid cell growth, then we would get uncontrolled dividing cells- called cancer cells. Thus, cancer is the consequence of at least two independent events- mutation of regulatory genes and mutation of growth-initiating genes. We speak of tumor initiation as first step and tumor progression as second step necessary for cancer formation. Of course there are coming more mutations in cell cycle control into game- but these mutations determine especially the behaviour of the cancer cell, i.e. if the tumor will be malign or not, or if it the tumor is growing fast or slow.
What does all this have to do with our Ecolicence to kill project?
What you do not know about cells until now is, that they secrete a wide range of different molecules to the environment. The interesting thing about cancer cells is that they secrete a different molecular pattern into the environment than normal, non-mutated cells. Even if cancer cells secrete similar molecules they pour them out in much higher or lower concentrations than normal cells just because they follow other metabolic paths (optimized for growth and cell division). We assume that it should be possible to distinguish normal cells and cancer cells by their molecular secretion-pattern. Our team built up a system taking advantage of this feature. In our proof of concept system for the iGEM competition we already constructed a small systems based on targeting cells by their molecular secretion pattern- as you already read. In accessory work outside of our project we will try to engineer bacteria able to recognize the molecular pattern of tumor cells and swimming in direction of the concentration of the secreted molecules. When arrived in the tumor’s environment the killing module of the cells should be activated and kill the cancer cells selectively and efficient. For the targeting we want to construct different chimeric chemotaxis receptors (as we did with LuxQ/Tar in our Ecolicence to kill project) and thus make the cells sensitive for tumor markers. We are aware of the fact that we will in many cases need more than just one marker being detected by our killer cells- the more molecular markers we detect, the more selective the targeting will be. Until now we are still far away from the medical application, but our iGEM project indicated that the idea of targeting and killing tumor cells is not so far-fatched. And we will hopefully follow up this approach in further research- with the motivation to one be able help many cancer patients in the world in the back of our heads. [back]
CONGRATULATIONS! You are really a very bright and interested fellow! And you really have a very good knowledge of synthetic biology and of the project of our team now, don't you? If you like a challenge, you can as a last step proceed to the scientific project description, starting with the sensing team ... ...final step
It was a pleasure for me to guide you around the Heidelberg iGEM Team and their Project on Synthetic biology. If you have further questions- just contact one of my scientist friends.
To get a better idea of real lab work visit our Lab impressions Photogalery.
And now: Have fun with the iGEM Project Heidelberg!!
[ ... back to step 5 ]
 "
"