Team:Heidelberg/Notebook/Killing II/7thweek
From 2008.igem.org
(Difference between revisions)
(New page: <html> <link rel='stylesheet' href='http://igem-heidelberg.de/fileadmin/Wiki/Heidelberg2.css' type='text/css' /> <link rel="stylesheet" href="http://igem-heidelberg.de/fileadmin/Wiki/Menu....) |
(→HisTag cloning of Colicins for purification) |
||
| Line 470: | Line 470: | ||
2.0 µl PstI | 2.0 µl PstI | ||
5.0 µl BSA 10x | 5.0 µl BSA 10x | ||
| - | 16.0 µl H<sub>2</ | + | 16.0 µl H<sub>2</sub>O |
------- | ------- | ||
50.0 µl | 50.0 µl | ||
| Line 477: | Line 477: | ||
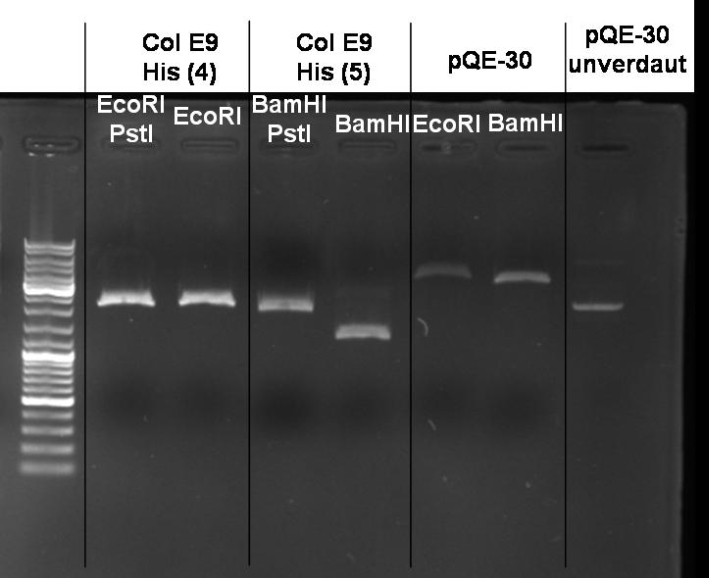
*Gel of Digestion: 1% Agarose,135 V, 40 min [[Image:080919-controldigestion_pQE-30_ColE9_small.jpg |500 px | center]] | *Gel of Digestion: 1% Agarose,135 V, 40 min [[Image:080919-controldigestion_pQE-30_ColE9_small.jpg |500 px | center]] | ||
*Gelresults: Maybe there is no insert or the enzymes didn't cut. | *Gelresults: Maybe there is no insert or the enzymes didn't cut. | ||
| + | |||
===Sender cloning: pBAD promoter=== | ===Sender cloning: pBAD promoter=== | ||
*BBa_I0500 ONC of 2008 stock did not grow | *BBa_I0500 ONC of 2008 stock did not grow | ||
Revision as of 08:43, 27 October 2008


7th week
Contents |
Monday 09/15/2008
pSB1A3-Receiver-Colicin cloning
- Colony-PCR-Screen of transformation for selection of positive clones: 108 colonies
25.0 µl Phusion Master Mix 2.5 µl T9002_lux_pR_rv_SpeI_BamHI_RBS 2.5 µl T9002_fw_XbaI 20.0 µl H2O 5 colonies ------- 50.0 µl
program:
98 °C 5 min 98 °C 10 sec | 58 °C 30 sec | 25 cycles 72 °C 45 sec | 72 °C 5 min 4 °C constant
- Gel of Colony-PCR-Screen: 1% Agarose, 135 V, 30 min
HisTag cloning of Colicins for purification
- Results of Transformation: Some colonies were grown on the LB-Ampicilin plates. Possibly some positive clones. Verification by Colony-PCR-Screen.
- Colony-PCR-Screen of transformation for selection of positive clones: 12 x colE1, 12 x colE9
25.0 µl Phusion Master Mix 2.5 µl ColE1_prot_fw_BamHI/ColE9_prot_fw_BamHI 2.5 µl ColE1_prot_rv_XmaI/ColE9_prot_rv_HindIII 20.0 µl H2O 5 colonies ------- 50.0 µl
program:
98 °C 5 min 98 °C 10 sec | 59 °C 30 sec | 25 cycles 72 °C 45 sec | 72 °C 5 min 4 °C constant
- Gel of Colony-PCR-Screen: 1% Agarose, 135 V, 30 min
- Colony-PCR-Screen of colony 1-10 colE9His to identify the positive clone
25.0 µl Phusion Master Mix 2.5 µl ColE1_prot_fw_BamHI/ColE9_prot_fw_BamHI 2.5 µl ColE1_prot_rv_XmaI/ColE9_prot_rv_HindIII 20.0 µl H2O 1 colony ------- 50.0 µl
program:
98 °C 5 min 98 °C 10 sec | 68 °C 30 sec | 25 cycles 72 °C 45 sec | 72 °C 5 min 4 °C constant
- Colony-PCR-Screen of colony 1-10 colE9His to identify the positive clone
25.0 µl Phusion Master Mix 2.5 µl ColE1_prot_fw_BamHI/ColE9_prot_fw_BamHI 2.5 µl ColE1_prot_rv_XmaI/ColE9_prot_rv_HindIII 20.0 µl H2O 1 colony ------- 50.0 µl
program:
98 °C 5 min 98 °C 10 sec | 68 °C 30 sec | 25 cycles 72 °C 45 sec | 72 °C 5 min 4 °C constant
Tuesday 09/16/2008
pSB1A3-Receiver-Colicin cloning
- Miniprep of ONC: eluted in 30 µl H2O (Qiagen, Miniprepkit)
- Controldigestion of Minipreps: 2 h -> 37 °C, 15 min -> 65 °C
10.0 µl DNA 3.0 µl H2O 2.0 µl NEBuffer I (NEB) 1.5 µl XbaI (20 000 U/ml, NEB) 1.5 µl SpeI (10 000 U/ml, NEB) 2.0 µl BSA 10x (NEB) ------- 20.0 µl
- Gel of digestion: 1% Agarose, 37 min, 135V
- Gelresults:
- Expected Fragments:
- pSB1A3 ~2000 bp
- T9002_without_GFP ~1088 bp
- The expected fragments were not visible. maybe the cloning was not succesfull.
- Expected Fragments:
- Digestion of Miniprpes with SpeI only for further cloninf, because double digestion with BamHI is not recommed. 2 h -> 37 °C, 10 min -> 65 °C
- Gel of Digestion: 1% Agarose, 30 min, 135 V
- PCR of Minipreps of "positive" clones: To check if we have positive clones we make PCR of the Minipreps we made.
25.0 µl Phusion MasterMix (Finnzymes, NEB) 2.5 µl Primer fw (T9002_Lux_pR_SpeI_BamHI_RBS) 2.5 µl Primer rv (T9002_fw_XbaI) 18.0 µl H2O 2.0 µl DNA-Template ------- 50.0 µl
program: 98 °C 30 sec 98 °C 10 sec | 58 °C 30 sec | 25 cycles 72 °C 45 sec | 72 °C 5 min 4 °C constant
- Gel of PCR products: 1% Agarose, 135 V, 30 min
- Gelresults:
- Expected Fragments:
- T9002_without_GFP ~1088 bp
- In each probe there is a fragment of ~1500 bp. We expect that this is a form of undigested plasmid. That means that if the PCR worked properly we have no positive clones. To recheck the Minipreps we try an additional PCR with our old protocoll (no Phusion MasterMix).
- Expected Fragments:
- PCR of Minipreps of "positive" clones: To check if we have positive clones we make PCR of the Minipreps we made.
10.0 µl 5x Phusion HF Buffer (Finnzymes, NEB) 1.0 µl 10 mM dNTPs (Invitrogen) 2.5 µl Primer fw (T9002_Lux_pR_SpeI_BamHI_RBS) 2.5 µl Primer rv (T9002_fw_XbaI) 31.5 µl H2O 2.0 µl DNA-Template 0.5 µl Phusion DNA-Polymerase (Finnzymes, NEB) ------- 50.0 µl
program: 98 °C 30 sec 98 °C 10 sec | 59 °C 30 sec | 25 cycles 72 °C 45 sec | 72 °C 5 min 4 °C constant
HisTag cloning of Colicins for purification
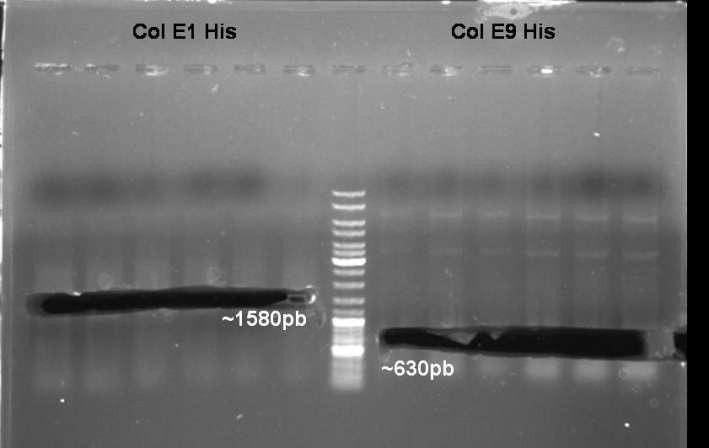
- Digestion of Minipreps with BamHI/HindIII for Colicin E1 and BamHI/XmaI for ColicinE9His: We want to select the positivt clones.
- Gel of Digestion: 1% Agarose, 135 V, 30 min
- Gelresults:
- Expected Fragments ColE1:
- ~1580 bp (Insert)
- ~3400 bp (Backbone)
- Expected Fragments ColE9:
- ~1580 bp (Insert)
- ~3400 bp (Backbone)
- None of the inserts is there. Maybe we have only religated backbone. Or maybe only one enzyme cutted the DNA.
- Expected Fragments ColE1:
- PCR of Minipreps to check if there are any positive clones. Programm see pSB1A2-Receiver-Cloning. Primers:
- Colicin E1:
- ColE1_prot_fw_BamHI
- ColE1_kil_prot_rv_SpeI
- Colicin E9:
- ColE9_prot_fw_BamHI
- ColE9_prot_rv_HindIII
- Colicin E1:
- Gel of ColicinE1 PCR: 1% Agarose, 135V, 30 min
- Gel of ColicinE9 PCR: 1% Agarose, 135V, 30 min
- Gelresults of PCR: There were no psitive bands for each PCR. But we used the wrong reverse primers. Because of that we started a new PCR with the right reverse primers (see protocol at pSB1A3-Receiver cloning)
Wednesday 09/17/2008
pSB1A3-Receiver-Colicin cloning
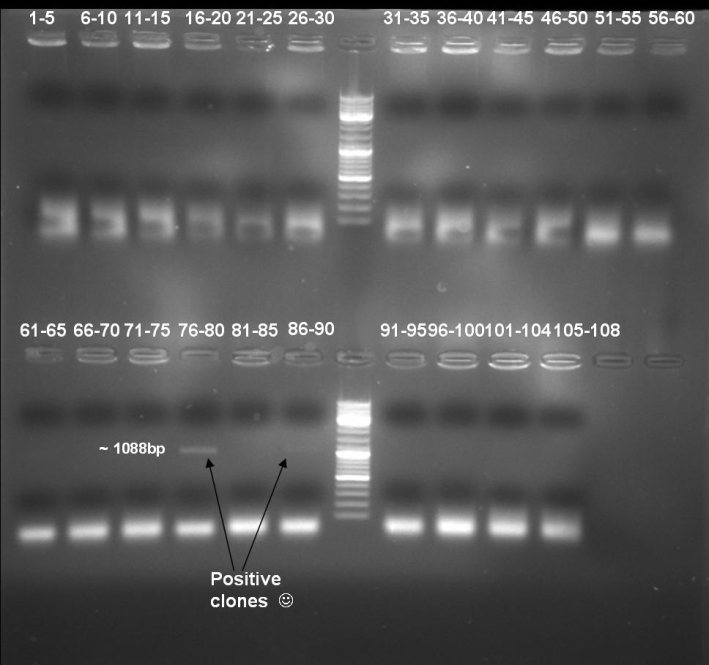
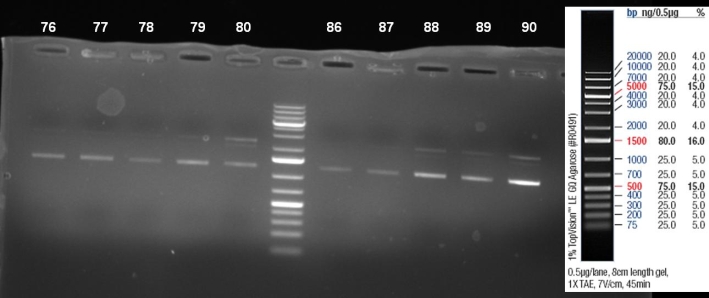
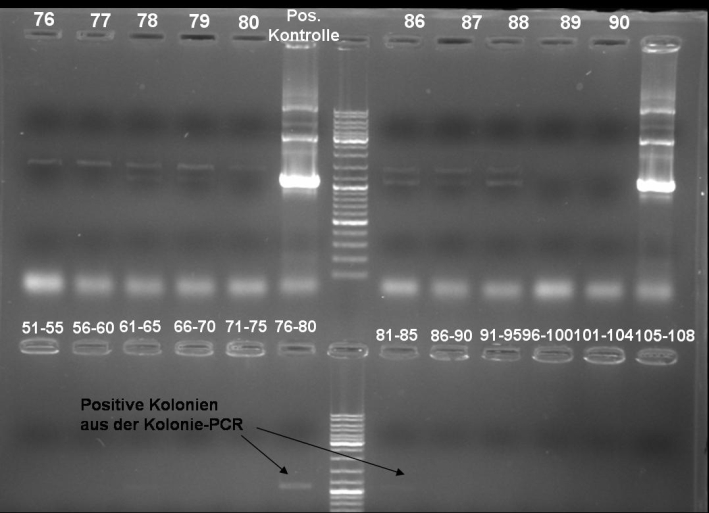
- Kontrolgel of PCR (09/16/2008) to select positive clones.
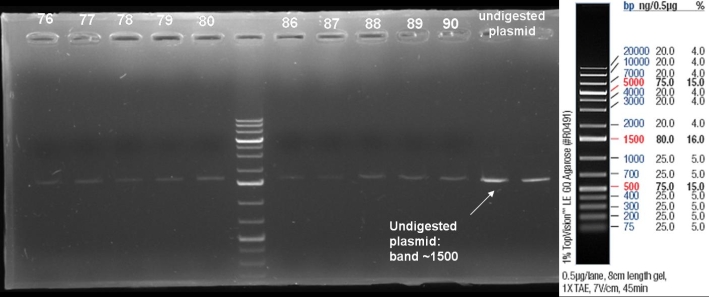
- Gelresults: positive clones should contain fragments at 1088 bp. Only the colonies 76-80 & 81 - 85 have a dimm band with this size. This could be a positive clone.
- Minipreps of these ten cultures: eluted in 30 µl H2O, Qiagen Miniprepkit
- Send plasmid DNA of colonies 78, 86, 87 & 88 to GATC for sequencing:
- Inoculation of ONC with Reference_promoter cells (BBa_I20260), Colicin E1 cells, Colicin E9 cells and MG1655 in 10 ml TB media.
HisTag cloning of Colicins for purification
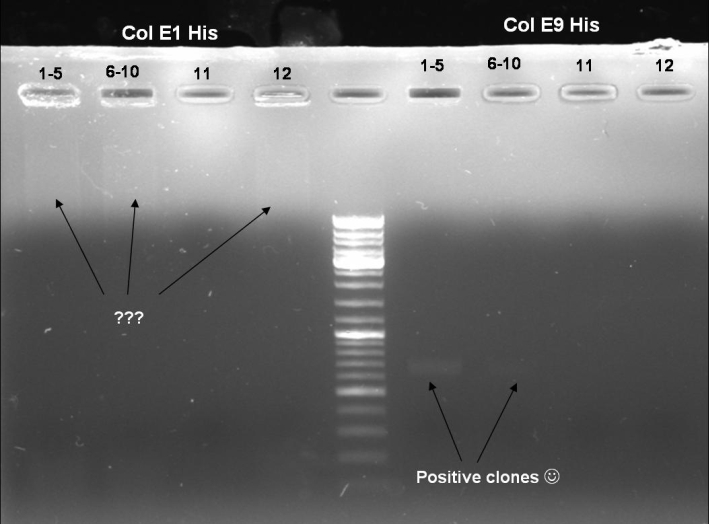
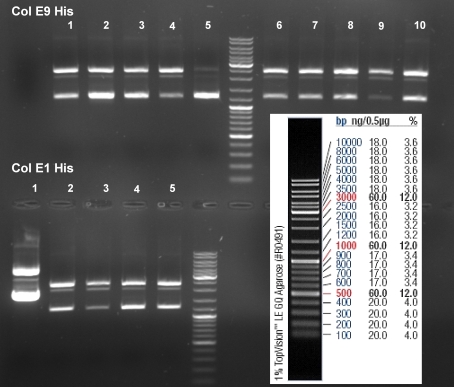
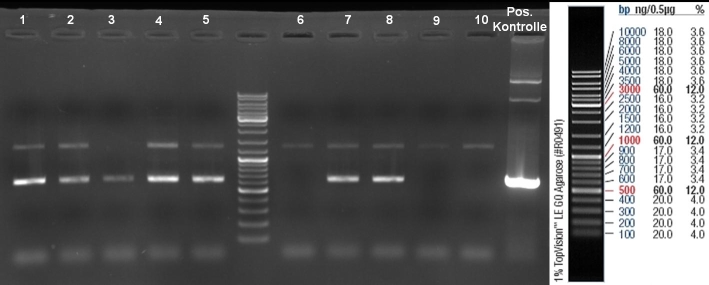
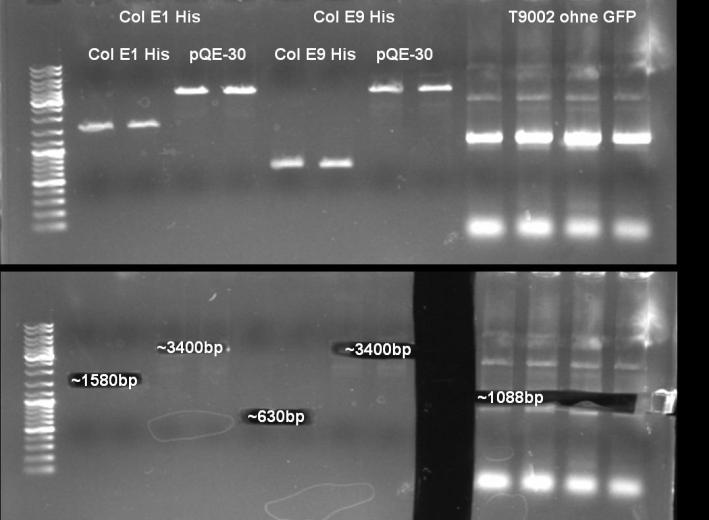
- Gel of ColE9 (top) ColE1 (bottom left) PCR screen (09/15/2008):1% Agarose, 30 min, 135 V
- Gel of ColE1 and ColE9 His PCR screen (09/15/2008): 1% Agarose, 30 min, 135 V
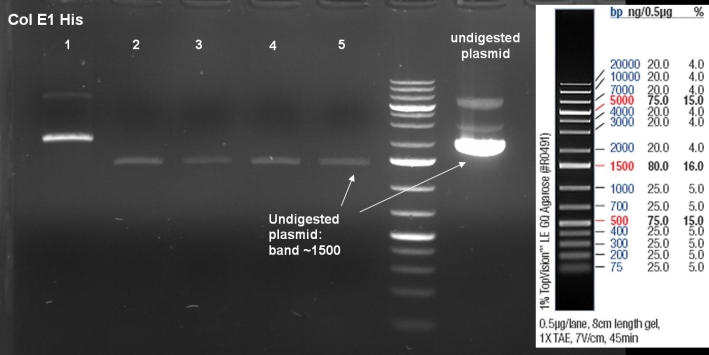
- Gel of ColE1 His PCR screen (09/15/2008): 1% Agarose, 30 min, 135 V
- Gelresults: Only dimm bands with the right fragment size. This could be the insert or a supercoiled form of the plasmid.
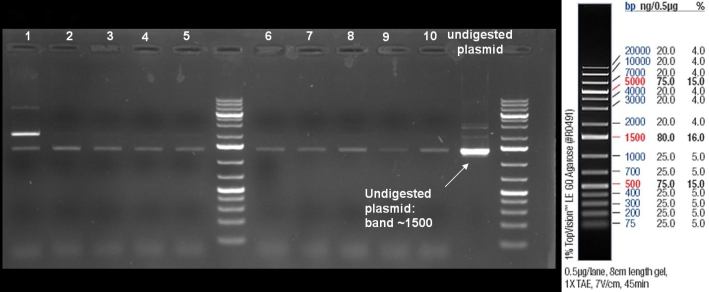
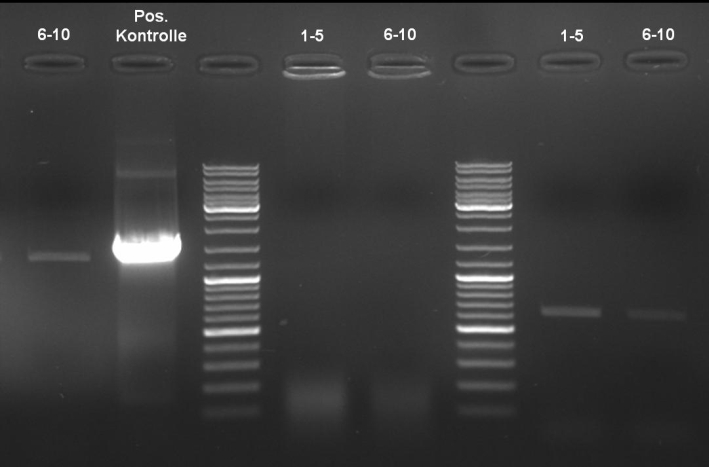
- ColE9 His PCR-Screen for selection of positive clones: 1% Agarose , 135 V, 30 min, expected fragment size ~630 bp
- Gelrsults: probes 1, 2, 3, 4, 5, 7 & 8 have bands with the right fragment size.
- Miniprepes of liquid ONC ColE9 His cultures 1-10 ColE1 His cultures 6-10: 1% Agarose, 135 V, 30 min
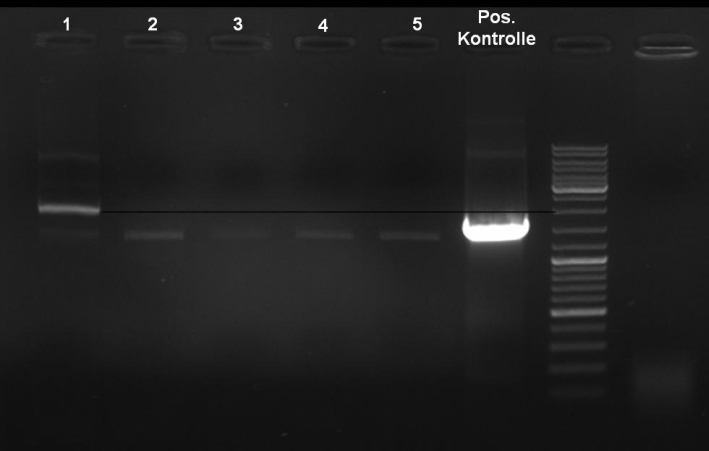
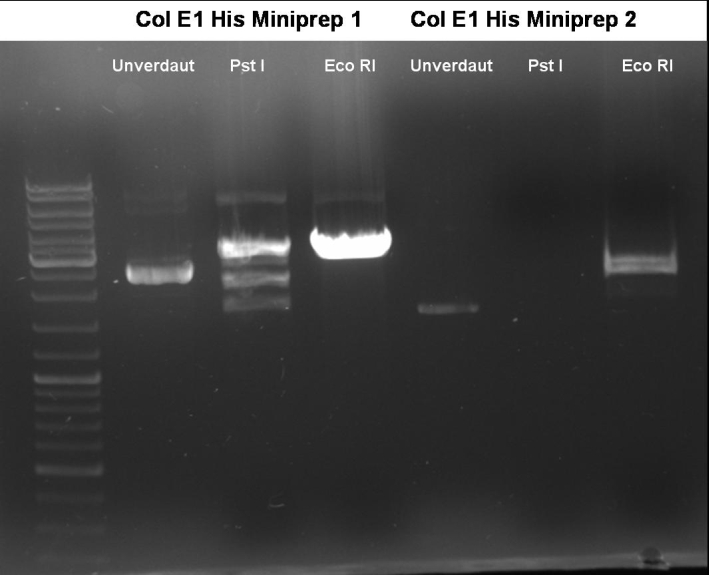
- Controldigestion of ColE1His 1 & 2 with EcoRI & PstI: 1% Agarose, 135 V, 30 min
- Results: fragments are to small. Fragments have the size of the pQE30-vector. -> Religated vector.
- Probes of ColE9 His cloning were send to GATC for sequencing.
Thursday 09/18/2008
pSB1A2-Receiver-Colicin cloning
- Sequencing results:
- only results from forward primer
- sequence is from backbone -> cloning was not succesfull
- maybe the backbone we used was already religated
- ->new start of cloning
- PCR of T9002 without GFP:
2.5 µl Primer reverse (T9002_Lux_pR_SpeI_BamHI_RBS) 2.5 µl Primer forward (T9002_fw_XbaI) 25.0 µl Phusion MM 18.0 µl H2O 2.0 µl Template DNA ------- 50.0 µl
program: 98 °C 30 sec 98 °C 10 sec | 58 °C 30 sec | 30 cycles 72 °C 45 sec | 72 °C 5 min 4 °C constant
- Gelextraction of PCR: 0.7% Agarose, 45 min, 135 V
- Digestion of Insert & Backbone: 1 h -> 37 °C, 10 min -> 65 °C
Insert 37.0 µl DNA of Gelex 2.0 µl SpeI 1.0 µl XbaI 5.0 µl NEBuffer 2 10x 5.0 µl BSA 10x ------- 50.0 µl
Backbone 37.0 µl T9002 Maxi (1 µg/µl) 2.0 µl SpeI 1.0 µl XbaI 5.0 µl NEBuffer 2 10x 5.0 µl BSA 10x ------- 50.0 µl
- Gelextraction of pSB1A3: 0.7% Agarose, 65 min, 100 V, eluated in 50 µl H2O
- Sapping of pSB1A3 backbone:
34 µl DNA 1 µl SAP-Enzymes (Fermentas) 4 µl SAP-Buffer (Fermentas) ----- 39 µl
- ON-Ligation: 14 h -> 16 °C, 20 min -> 65 °C, forever -> 4 °C
5:1 Insert:Vector 15.0 µl Insert 1.0 µl Vector 2.0 µl T4 DNA Ligase 2.0 µl T4 DNA Ligase Buffer ------- 20.0 µl
3:1 Insert:Vector 14.5 µl Insert 1.5 µl Vector 2.0 µl T4 DNA Ligase 2.0 µl T4 DNA Ligase Buffer ------- 20.0 µl
Colicin-Test with eucaryotic cells
- HeLa and MCF7 cells were treated with supernatants of prior tests. There was no effect in the first hour. Maybe the supernatant was uneffective because it was freezed.
HisTag cloning of Colicins for purification
- Sequencing results:
- religated pQE-30 -> no positive clones
- ->restart the cloning
- PCR of colicin E1/E9 proteins:
2.0 µl pColE1/pColE9-J plasmid 2.5 µl Primer fw(ColE1_prot_fw_BamHI/ColE9_prot_fw_BamHI) 2.5 µl Primer rv(ColE1_prot_rv_HindIII/ColE9_prot_rv_XmaI) 18.0 µl H2O 25.0 µl Phusion MM ------- 50.0 µl
- Gelextraction of PCR: 0,7% Agarose, 50 min, 100 V; eluted in 30 µl H2O, Qiagen Gelextraction Kit
- 1st Digestion of Vectors and inserts with HindIII/XmaI for ColE1/E9 HisTag cloning: 1h -> 37 °C, 10 min -> 65 °C
vector: 39.8 µl pQE30 (30 ng/µl) 0.2 µl HindIII/XmaI 5.0 µl NEBuffer 2/4 10x 5.0 µl BSA 10x ------- 50.0 µl
insert: 10.0 µl colicinE1/E9 0.2 µl HindIII/XmaI 5.0 µl NEBuffer 2/4 10x 5.0 µl BSA 10x 29.8 µl H2O ------- 50.0 µl
- Purification of probes with Qiagen PCR Purification Kit
- 2nd Digestion of Vectors and inserts with BamHIfor ColE1/E9 HisTag cloning: 1h -> 37 °C, 10 min -> 65 °C
39.0 µl pQE30 (30 ng/µl) 1.0 µl BamHI 5.0 µl NEBuffer 3 10x 5.0 µl BSA 10x ------- 50.0 µl
insert: 39.0 µl colicinE1/E9 1.0 µl BamHI 5.0 µl NEBuffer 2/4 10x 5.0 µl BSA 10x 29.8 µl H2O ------- 50.0 µl
- Gelextraction of digested vector and insert: 0.7% Agarose, 45 min, 100 V
- ON Ligation of pQE-30 vector and colicin inserts: 14 h -> 16 °C, 20 min -> 65 °C, forever -> 4 °C
12.0 µl Insert 4.0 µl Vector 2.0 µl T4 DNA Ligase 2.0 µl T4 DNA Ligase Buffer ------- 20.0 µl
Other
- Inoculation of ONC with
- BBa_I23107 -> medium constitutive promoter
- BBa_I13600 -> CFP cassette
- BBa_I13602 -> CFP cassette
- BBa_B0015 -> Terminator
- BBa_I0500 -> pBAD/araC
- BBa_F1610 -> AHL sender part
Friday 09/19/2008
pSB1A3-Receiver-Colicin cloning
- Transformation of pSB1A3-T9002-without-GFP ON-Ligations (Transformation protocol Chris)
2x 5 µl DNA 5:1 on 50 µl TOP 10 cells + 200 ml LB -> LB-Amp plates 2x 5 µl DNA 3:1 on 50 µl TOP 10 cells + 200 ml LB -> LB-Amp plates
HisTag cloning of Colicins for purification
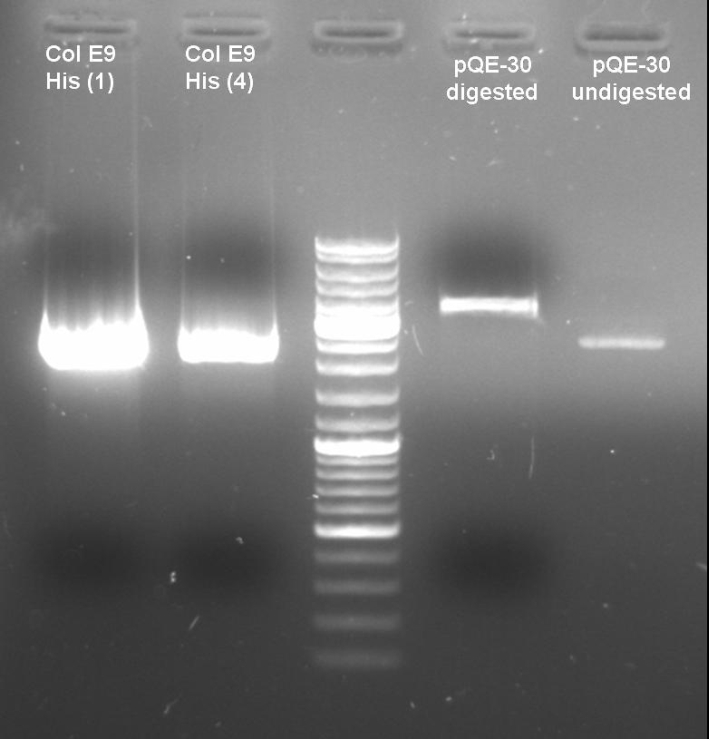
- Digestion of old ColE9His-pQE-30 cloning with BamHI/PstI to verify that there are no positive clones: colonies 1 + 4; 1h 10min -> 37 °C
20.0 µl DNA 5.0 µl NEBuffer 3 2.0 µl BamHI 2.0 µl PstI 5.0 µl BSA 10x 16.0 µl H2O ------- 50.0 µl
- Transformation of ColE9/E1 His tag cloning ON-Ligations (Transformation protocol Chris)
2x 5 µl DNA 5:1 on 50 µl TOP 10 cells + 200 ml LB -> LB-Amp plates
- Gel of Digestion: 1% Agarose,135 V, 40 min
- Gelresults: Maybe there is no insert or the enzymes didn't cut.
Sender cloning: pBAD promoter
- BBa_I0500 ONC of 2008 stock did not grow
- Inoculation of BBa_I0500 of Glycerolstock 2008 & 2004
Other
Team-Meeting
- Presentationpreparation for Team-Meeting: Which parts can be submitted to the registry?
- Presentation:
- ColicinE1 and E9 as part whole cassette
- Mutagenesis of ColicinE1 Receiver (EcoRI)
Minipreps
- Of ONCs, eluted in 30µl H2; Qiagen Miniprepkit
Saturday 09/20/2008
pSB1A3-Receiver-Colicin cloning
- Colony PCR-Screen of pSB1A2-T9002-GFP:
25.0 µl Phusion MasterMix (Finnzymes, NEB) 2.5 µl Primer_fw (pSB_ins_fw) 2.5 µl Primer_rv (pSB_ins_rv) 20.0 µl H2O 5 colonies ------- 50.0 µl
program: 98 °C 5 min 98 °C 10 sec | 58 °C 30 sec | 25 cycles 72 °C 45 sec | 72 °C 5 min 4 °C constant
- Gel of PCR screen: No bands were visible. Retry PCR-Screen with Taq-Polymerase and other primers.
HisTag cloning of Colicins for purification
- Digestion of former Cloning with ColE9His-pQE30 EcoRI/PstI and BamHI/PstI: 1h 37 °C -> Gel
20.0 µl DNA 5.0 µl NEBuffer 3 2.0 µl BamHI 2.0 µl PstI 5.0 µl BSA 10x 16.0 µl H2O ------- 50.0 µl
20.0 µl DNA 5.0 µl NEBuffer EcoRI 2.0 µl EcoRI 2.0 µl PstI 5.0 µl BSA 10x 16.0 µl H2O ------- 50.0 µl
20.0 µl DNA 5.0 µl NEBuffer 3 2.0 µl BamHI 5.0 µl BSA 10x 18.0 µl H2O ------- 50.0 µl
20.0 µl DNA 5.0 µl NEBuffer EcoRI 2.0 µl EcoRI 5.0 µl BSA 10x 18.0 µl H2O ------- 50.0 µl
- Gel of digestion: 1% Agarose, 135V, 30 min:
- Inoculation of ONC of 25 colonies per colicin-pQE30
Sunday 09/21/2008
pSB1A3-Receiver-Colicin cloning
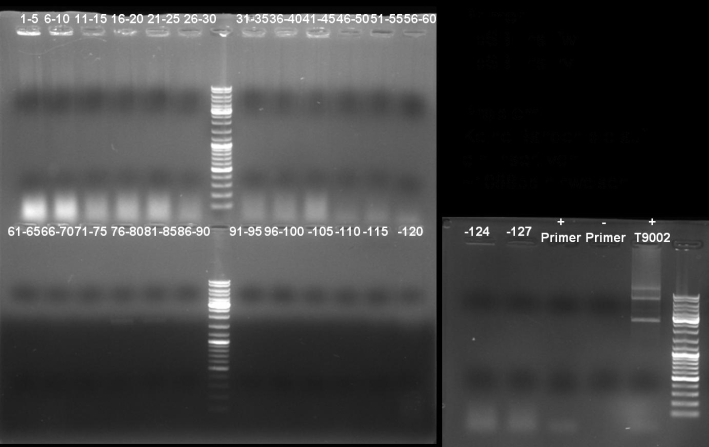
- 2nd PCR Screening of pSB1A2-T9002-GFP cloning
25.0 µl Taq-Polymerase MasterMix (Fermentas) 2.5 µl Primer_rv (T9002_Lux_pR_SpeI_BamHI_RBS) 2.5 µl Primer_fw (T9002_fw_XbaI) 20.0 µl H2O 5 colonies ------- 50.0 µl
program: 95 °C 3 min 95 °C 1 min | 54 °C 1 min | 30 cycles 72 °C 1 min | 72 °C 10 min 4 °C constant
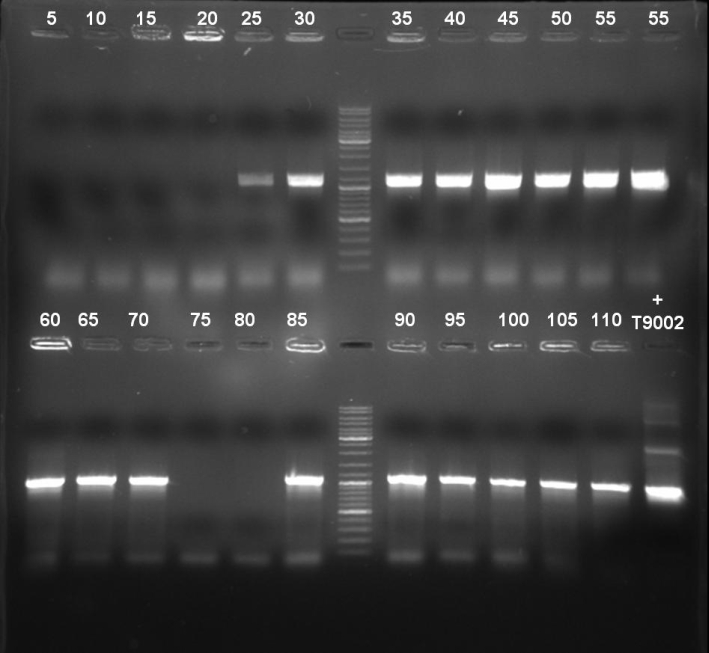
- Gel of colony PCR screen: 1% Agarose, 135 V, 30 min
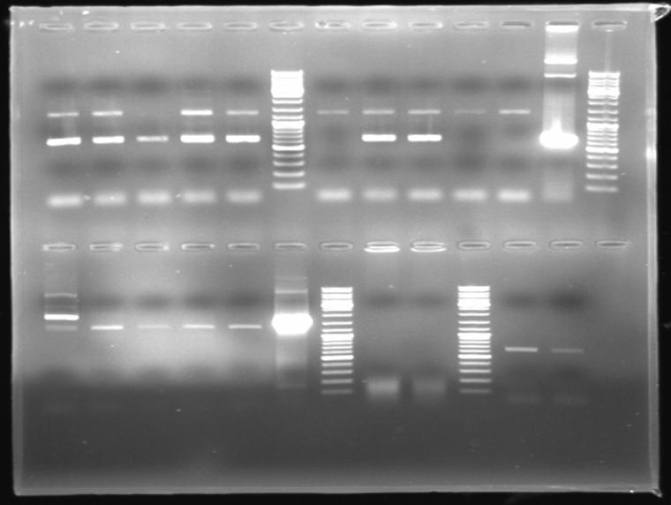
- Gelresults: Many bands with the right fragment size. New colony PCR screen with single colonies of colonies 31-45 and 66-75
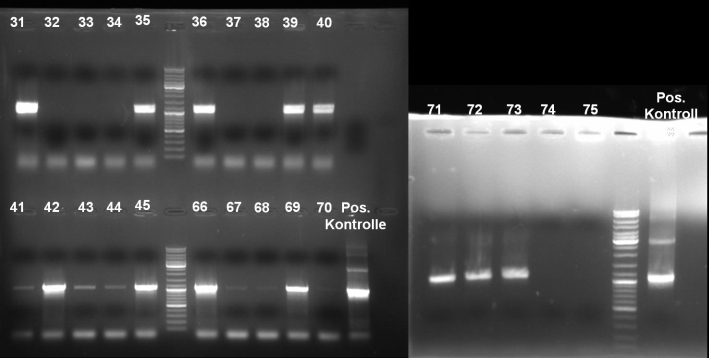
- Gel of single colony PCR screen: 1% Agarose, 135 V, 30 min
- Gelresults: Colonies 31, 35, 36, 39, 40, 42, 45, 66, 69, 71, 72 & 73 have the right fragment size. This could be the positive clones
- Inoculation of LB-Amp ONC from these colonies
HisTag cloning of Colicins for purification
- Colony PCR screen to select positive clones:
25.0 µl Taq-Polymerase MasterMix (Fermentas) 2.5 µl Primer_rv 2.5 µl Primer_fw 20.0 µl H2O 5 colonies ------- 50.0 µl
program: 95 °C 3 min 95 °C 1 min | 54 °C 1 min | 30 cycles 72 °C 1 min | 72 °C 10 min 4 °C constant
 "
"