Team:Illinois/Bimolecular Fluorescence Biosensor
From 2008.igem.org
(→Literature References) |
|||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:bimolflour.png|center]] | |
| - | + | {{bottom_template}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Core Team Members== | ==Core Team Members== | ||
| - | Luke Edelman, Adam Zoellner, Meghan McCleary, Katrina Keller | + | [[User:Lbedelman|Luke Edelman]], [[User:azoelln2|Adam Zoellner]], [[User:MMcCleary|Meghan McCleary]], [[User:Trina333|Katrina Keller]] |
| - | ==Project | + | ==Project Abstract== |
We hope to design a soluble molecular biosensor that, when it comes in contact with an 'activating' ligand such as a virus, bacterium, or specific antibody, generates a fluorescent response using bimolecular complementation. Traditionally unimolecular constructs such as Green Fluorescent Protein (GFP) can be split into two heterologous protein fragments, which can then bind and reinitiate fluorescence upon close spatial proximity. GFP fragments are often fused to endogenous intracellular proteins to study protein-protein interactions: complementation between these GFP fragments is achieved only when they are tethered to proteins which interact (bind) strongly. | We hope to design a soluble molecular biosensor that, when it comes in contact with an 'activating' ligand such as a virus, bacterium, or specific antibody, generates a fluorescent response using bimolecular complementation. Traditionally unimolecular constructs such as Green Fluorescent Protein (GFP) can be split into two heterologous protein fragments, which can then bind and reinitiate fluorescence upon close spatial proximity. GFP fragments are often fused to endogenous intracellular proteins to study protein-protein interactions: complementation between these GFP fragments is achieved only when they are tethered to proteins which interact (bind) strongly. | ||
| - | We seek to harness this powerful molecular tool for an inverse task: instead of studying putative interactions between known proteins, we are designing fusion constructs to detect the presence of proteins known ''a priori'' to interact, for deployment as a one-step diagnostic assay. For example, virus envelopes are often composed of large multiprotein complexes; one GFP fragment could be fused to an antibody against | + | We seek to harness this powerful molecular tool for an inverse task: instead of studying putative interactions between known proteins, we are designing fusion constructs to detect the presence of proteins known ''a priori'' to interact, for deployment as a one-step diagnostic assay. For example, virus envelopes are often composed of large multiprotein complexes; one GFP fragment could be fused to an antibody against a particular envelope protein, and the complementary fragment fused to an antibody against an adjacent protein. In solution, only the presence of this specific multiprotein complex would generate GFP complementation and the resulting fluorescence, providing a robust one-step method for the detection of biological pathogens. Similarly, many secreted bacterial enterotoxins form multiprotein complexes, such as cholera toxin. Additionally, many immunoglobulin biomolecules can form aggregates in the circulation. |
| + | |||
| + | ==Specific Plans, Supplies, and Protocols== | ||
| + | |||
| + | We are currently considering possible biological targets for use as an initial proof-of-concept. Specifically, this project requires a known protein-protein interaction: for example, a ligand-receptor pair, or an antibody-epitope pair, for each of two unique sites on a target pathogenic protein. | ||
| + | |||
| + | One possibility we are currently considering is an antibody to the cholera toxin B subunit (CTB) which forms a homopentameric complex in solution and can be purchased commercially. In this case, we would fuse both GFP fragments to an antibody against CTB. Fluorescence would occur when a CTB complex exists to tether two complementary GFP fragments through the CTB-antibody interaction. | ||
| + | |||
| + | A second possible case study is antibody-ligand interactions: we could fuse GFP fragments to known immunogenic proteins or protein fragments from viruses or bacteria. Since immunoglobulin molecules are intrinsically bivalent in their ligand interactions, this provides a natural tethering to enable complementation. | ||
| + | |||
| + | Once we decide on an initial ligand pairing, we must acquire genes for our desired proteins- both the GFP fragments, and the fusion ligand, as well as the target protein or antibody if it cannot be purchased commercially. | ||
| + | |||
| + | We must then design the appropriate primers and PCR procedure to enable this fusion, with an appropriate amino acid spacer between the two functional domains. Once this experimental procedure has been completed, we will evaluate the functionality of our end products on known concentrations of our target biomolecule. | ||
==Literature References== | ==Literature References== | ||
| Line 30: | Line 34: | ||
[https://static.igem.org/mediawiki/2008/f/fa/Yeast_BiFC_Plasmid_Construction.pdf Yeast BiFC Plasmid Construction] | [https://static.igem.org/mediawiki/2008/f/fa/Yeast_BiFC_Plasmid_Construction.pdf Yeast BiFC Plasmid Construction] | ||
| - | ==Planned Labwork== | + | ==Planned Labwork, Procedures, and Results== |
| + | [[Image:HIV_PIC.png|400px|right]] | ||
| + | |||
| + | HIV is different in structure from other retroviruses. It is around 120 nm in diameter (120 billionths of a meter; around 60 times smaller than a red blood cell) and roughly spherical. | ||
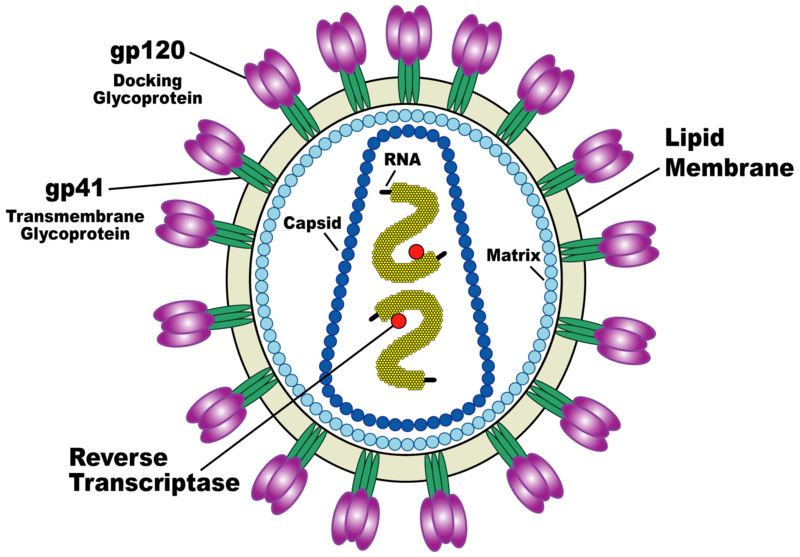
| + | HIV-1 is composed of two copies of single-stranded RNA enclosed by a conical capsid comprising the viral protein p24, typical of lentiviruses (Figure 1). The RNA component is 9749 nucleotides long[5]. This is in turn surrounded by a plasma membrane of host-cell origin. The single-strand RNA is tightly bound to the nucleocapsid proteins, p7 and enzymes that are indispensable for the development of the virion, such as reverse transcriptase and integrase. The nucleocapsid (p7 and p6) associates with the genomic RNA (one molecule per hexamer) and protects the RNA from digestion by nucleases. A matrix composed of an association of the viral protein p17 surrounds the capsid, ensuring the integrity of the virion particle. Also enclosed within the virion particle are Vif, Vpr, Nef, p7 and viral protease (Figure 1). The envelope is formed when the capsid buds from the host cell, taking some of the host-cell membrane with it. The envelope includes the glycoproteins gp120 and gp41. | ||
| + | In 2006, an Anglo-German team compiled a 3D structure of HIV by combining multiple images. It is hoped that this new information would contribute to scientific understanding of the virus, and help in the creation of a cure. Oxford University's Professor Stephen D. Fuller said the 3D map would assist in understanding how the virus grows. [6] The validity of this work remains a matter of debate [7], with a conflicting model produced by another team led by Florida State University Professor Kenneth Roux in the US [8]. (http://en.wikipedia.org/wiki/Structure_and_genome_of_HIV#gp120) | ||
| + | |||
| + | {{bottom_template}} | ||
Latest revision as of 07:17, 18 October 2008
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
Contents |
Core Team Members
Luke Edelman, Adam Zoellner, Meghan McCleary, Katrina Keller
Project Abstract
We hope to design a soluble molecular biosensor that, when it comes in contact with an 'activating' ligand such as a virus, bacterium, or specific antibody, generates a fluorescent response using bimolecular complementation. Traditionally unimolecular constructs such as Green Fluorescent Protein (GFP) can be split into two heterologous protein fragments, which can then bind and reinitiate fluorescence upon close spatial proximity. GFP fragments are often fused to endogenous intracellular proteins to study protein-protein interactions: complementation between these GFP fragments is achieved only when they are tethered to proteins which interact (bind) strongly.
We seek to harness this powerful molecular tool for an inverse task: instead of studying putative interactions between known proteins, we are designing fusion constructs to detect the presence of proteins known a priori to interact, for deployment as a one-step diagnostic assay. For example, virus envelopes are often composed of large multiprotein complexes; one GFP fragment could be fused to an antibody against a particular envelope protein, and the complementary fragment fused to an antibody against an adjacent protein. In solution, only the presence of this specific multiprotein complex would generate GFP complementation and the resulting fluorescence, providing a robust one-step method for the detection of biological pathogens. Similarly, many secreted bacterial enterotoxins form multiprotein complexes, such as cholera toxin. Additionally, many immunoglobulin biomolecules can form aggregates in the circulation.
Specific Plans, Supplies, and Protocols
We are currently considering possible biological targets for use as an initial proof-of-concept. Specifically, this project requires a known protein-protein interaction: for example, a ligand-receptor pair, or an antibody-epitope pair, for each of two unique sites on a target pathogenic protein.
One possibility we are currently considering is an antibody to the cholera toxin B subunit (CTB) which forms a homopentameric complex in solution and can be purchased commercially. In this case, we would fuse both GFP fragments to an antibody against CTB. Fluorescence would occur when a CTB complex exists to tether two complementary GFP fragments through the CTB-antibody interaction.
A second possible case study is antibody-ligand interactions: we could fuse GFP fragments to known immunogenic proteins or protein fragments from viruses or bacteria. Since immunoglobulin molecules are intrinsically bivalent in their ligand interactions, this provides a natural tethering to enable complementation.
Once we decide on an initial ligand pairing, we must acquire genes for our desired proteins- both the GFP fragments, and the fusion ligand, as well as the target protein or antibody if it cannot be purchased commercially.
We must then design the appropriate primers and PCR procedure to enable this fusion, with an appropriate amino acid spacer between the two functional domains. Once this experimental procedure has been completed, we will evaluate the functionality of our end products on known concentrations of our target biomolecule.
Literature References
Bimolecular Fluorescence Complementation Review
Yeast BiFC Plasmid Construction
Planned Labwork, Procedures, and Results
HIV is different in structure from other retroviruses. It is around 120 nm in diameter (120 billionths of a meter; around 60 times smaller than a red blood cell) and roughly spherical. HIV-1 is composed of two copies of single-stranded RNA enclosed by a conical capsid comprising the viral protein p24, typical of lentiviruses (Figure 1). The RNA component is 9749 nucleotides long[5]. This is in turn surrounded by a plasma membrane of host-cell origin. The single-strand RNA is tightly bound to the nucleocapsid proteins, p7 and enzymes that are indispensable for the development of the virion, such as reverse transcriptase and integrase. The nucleocapsid (p7 and p6) associates with the genomic RNA (one molecule per hexamer) and protects the RNA from digestion by nucleases. A matrix composed of an association of the viral protein p17 surrounds the capsid, ensuring the integrity of the virion particle. Also enclosed within the virion particle are Vif, Vpr, Nef, p7 and viral protease (Figure 1). The envelope is formed when the capsid buds from the host cell, taking some of the host-cell membrane with it. The envelope includes the glycoproteins gp120 and gp41. In 2006, an Anglo-German team compiled a 3D structure of HIV by combining multiple images. It is hoped that this new information would contribute to scientific understanding of the virus, and help in the creation of a cure. Oxford University's Professor Stephen D. Fuller said the 3D map would assist in understanding how the virus grows. [6] The validity of this work remains a matter of debate [7], with a conflicting model produced by another team led by Florida State University Professor Kenneth Roux in the US [8]. (http://en.wikipedia.org/wiki/Structure_and_genome_of_HIV#gp120)
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
 "
"