Team:Imperial College/B.subtilis
From 2008.igem.org
m |
m |
||
| (4 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
|[[Image:B.subtilis parts.PNG|right|300px]] | |[[Image:B.subtilis parts.PNG|right|300px]] | ||
}} | }} | ||
| - | {{Imperial/Box1|Transformation of ''B. subtilis''|In order to expand the use of ''B. subtilis'' as a potential chassis we investigated the different protocols used for the transformation of ''B. subtilis''. ''B. subtilis'' is used as the model for gram positive bacterium and have been extensively studied. A variety of protocols have been developed for the transformation of ''B. subtilis''. We investigated two different protocols, one relying upon natural competence of stressed ''B. subtilis'' and the other using electroporation. The protocols and results were both fully documented to further the use of ''B. subtilis'' as a chassis - The protocols can be found in the wet lab section (transformation 1 & 2). | + | {{Imperial/Box1|Transformation of ''B. subtilis''|In order to expand the use of ''B. subtilis'' as a potential chassis we investigated the different protocols used for the transformation of ''B. subtilis''. ''B. subtilis'' is used as the model for gram positive bacterium and have been extensively studied. A variety of protocols have been developed for the transformation of ''B. subtilis''. We investigated two different protocols, one relying upon natural competence of stressed ''B. subtilis'' and the other using electroporation. The protocols and results were both fully documented to further the use of ''B. subtilis'' as a chassis - The protocols can be found in the wet lab section (transformation 1 & 2), and you can click "Details" below to see the results of the transformation. |
| + | |||
| + | [[Team:Imperial_College/Transformation_Results |'''>>> Details >>>''']] | ||
| + | }} | ||
{{Imperial/Box1|Chassis Characterisation| | {{Imperial/Box1|Chassis Characterisation| | ||
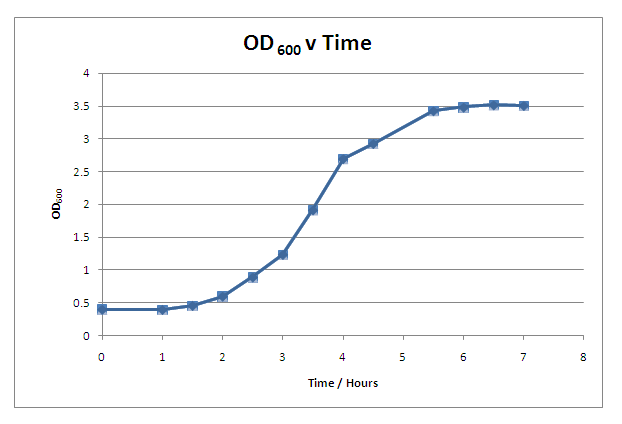
| - | + | ====== Growth Curve ====== | |
| - | We characterised the growth rate of ''B. subtilis'' in LB media at 37<sup>o</sup>C. This curve provides useful information when inducing protein synthesis in ''B. subtilis'' and when characterising parts such as promoters. In addition to collecting this growth curve information, we | + | We characterised the growth rate of ''B. subtilis'' in LB media at 37<sup>o</sup>C. This curve provides useful information when inducing protein synthesis in ''B. subtilis'' and when characterising parts such as promoters. In addition to collecting this growth curve information, we modelled the growth rate of ''B. subtilis'' which can be found in the [https://2008.igem.org/Team:Imperial_College/Growth_Curve Growth Curve] section of the Dry Lab. |
| - | [[Image:OD600 v Time.PNG|300px]] | + | ====== Motility ====== |
| + | We also went some way toward characterising the motility of the chassis but more detailed information on that side of the chassis characterisation can be found on the Major Results page and the motility section of the dry lab. | ||
| + | |[[Image:OD600 v Time.PNG|300px]] | ||
}} | }} | ||
{{Imperial/EndPage|Major_Results|Major_Results}} | {{Imperial/EndPage|Major_Results|Major_Results}} | ||
Latest revision as of 03:41, 30 October 2008
|
|||||||||||||||||||
 "
"