Team:Imperial College/B.subtilis
From 2008.igem.org
(New page: {{Imperial/StartPage2}} <br> {{Imperial/Box2|Working With ''B. subtilis''|Throughout this summer we worked with ''B. subtilis'' as a chassis. Although this chassis has great potential it i...) |
m |
||
| Line 10: | Line 10: | ||
}} | }} | ||
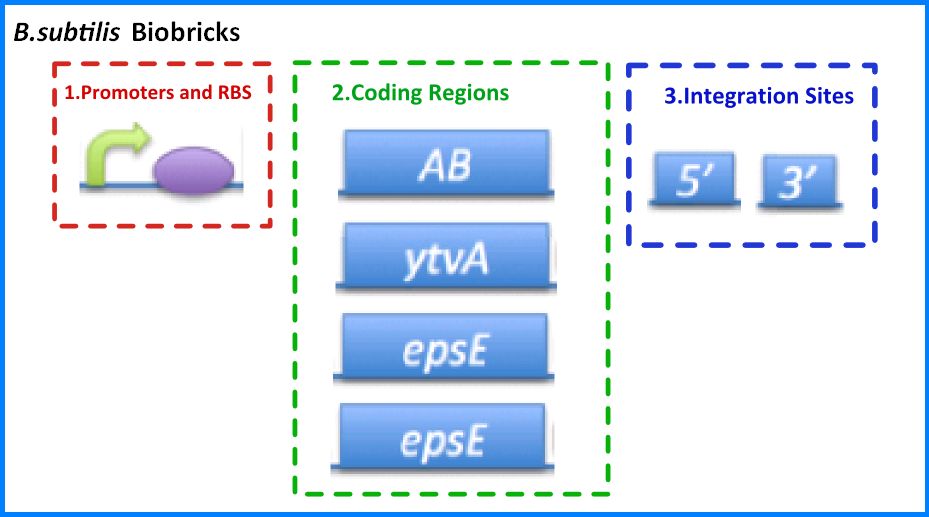
{{Imperial/Box1|Transformation of ''B. subtilis''|In order to expand the use of ''B. subtilis'' as a potential chassis we investigated the different protocols used for the transformation of ''B. subtilis''. ''B. subtilis'' is used as the model for gram positive bacterium and have been extensively studied. A variety of protocols have been developed for the transformation of ''B. subtilis''. We investigated two different protocols, one relying upon natural competence of stressed ''B. subtilis'' and the other using electroporation. The protocols and results were both fully documented to further the use of ''B. subtilis'' as a chassis. | {{Imperial/Box1|Transformation of ''B. subtilis''|In order to expand the use of ''B. subtilis'' as a potential chassis we investigated the different protocols used for the transformation of ''B. subtilis''. ''B. subtilis'' is used as the model for gram positive bacterium and have been extensively studied. A variety of protocols have been developed for the transformation of ''B. subtilis''. We investigated two different protocols, one relying upon natural competence of stressed ''B. subtilis'' and the other using electroporation. The protocols and results were both fully documented to further the use of ''B. subtilis'' as a chassis. | ||
| - | *The protocols can be found here -[https://2008.igem.org/Team:Imperial_College/Transformation Protocol 1] and [https://2008.igem.org/Team:Imperial_College/Transformation_2 Protocol 2] | + | *The protocols can be found here - [https://2008.igem.org/Team:Imperial_College/Transformation Protocol 1] and [https://2008.igem.org/Team:Imperial_College/Transformation_2 Protocol 2] |
| - | *The summary of results can be found here -[https://2008.igem.org/Team:Imperial_College/Transformation_Results Summary of results] | + | *The summary of results can be found here - [https://2008.igem.org/Team:Imperial_College/Transformation_Results Summary of results] |
}} | }} | ||
Revision as of 17:53, 29 October 2008
|
|||||||||||||||||||
 "
"