Team:Imperial College/Motility
From 2008.igem.org
m |
|||
| Line 2: | Line 2: | ||
=== Motility Analysis === | === Motility Analysis === | ||
{{Imperial/Box2|| | {{Imperial/Box2|| | ||
| - | |||
<br><br> | <br><br> | ||
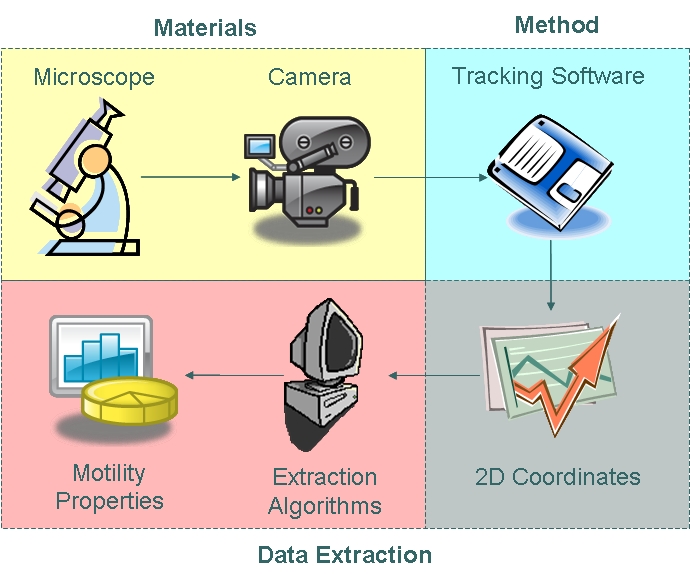
| - | + | As part of our chasis characterisation process, we have decided to model ''B. subtilis'' motility. In order to do this, the approach illustrated below was taken. The first phase of modelling involved data collection using microscopy techniques and cell tracking. Collected data was then analysed using algorithms which enabled us to extract distributions of parameters as defined in our model. | |
| + | |||
| + | [[Image:Approach.jpg|450px|center]] | ||
| + | |||
| + | ===== Materials ===== | ||
| + | |||
| + | We used the Zeiss Axiovert 200 inverted microscope and Improvision Volocity acquisition software. This system offers a full incubation chamber with temperature control and a highly sensitive 1300x1000 pixel camera for fast low-light imaging. Video images are captured into memory by the system at a basal video frame rate of 16.3Hz. This can be further increased to 27.9Hz by performing x4 binning. | ||
| + | |||
| + | |||
| + | ===== Method ===== | ||
| + | |||
| + | We used manual tracking to track ''B. subtilis'', obtaining two-dimensional coordinate data points which describes by the trajectory of the cells. | ||
| + | |||
| + | ===== Data Extraction ===== | ||
| + | |||
| + | We input the coordinate data into algorithms to model cell trajectory and motility. | ||
| + | |||
| + | === Motility Model === | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
}} | }} | ||
{{Imperial/EndPage|Genetic Circuit|Appendices}} | {{Imperial/EndPage|Genetic Circuit|Appendices}} | ||
Revision as of 08:49, 17 October 2008
Motility Analysis
|
|||||||
 "
"