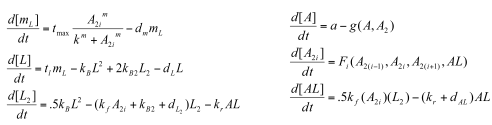Team:Michigan/Project/Modeling/Model2.html
From 2008.igem.org
(Difference between revisions)
| Line 10: | Line 10: | ||
While Model 1 gave us some important results (mainly: need a low Kd for oscillations to occur) , we decided to look at more complete model that accounted for some of the dimerizations the proteins undergo: | While Model 1 gave us some important results (mainly: need a low Kd for oscillations to occur) , we decided to look at more complete model that accounted for some of the dimerizations the proteins undergo: | ||
<div align='center'> [[Image: Model2Equations.png]]</div> | <div align='center'> [[Image: Model2Equations.png]]</div> | ||
| - | Parameters | + | Parameters |
*a= production of NifA | *a= production of NifA | ||
*kf= binding rate of NifA and NifL | *kf= binding rate of NifA and NifL | ||
| Line 17: | Line 17: | ||
*kb:rate of complex formation (i.e. dimerization, formation of tetramers/hexamers) | *kb:rate of complex formation (i.e. dimerization, formation of tetramers/hexamers) | ||
*kb2: dissociation rate of such complexes given above | *kb2: dissociation rate of such complexes given above | ||
| + | *m= represents how many subunits of NifA bind to NifHp (i.e., if we modeled NifA as a dimer [so 'i'=2], we would say m=3, since 2*3=6 (NifA is a hexamer). | ||
| + | *tmax: Maximal transcription rate | ||
| + | *tl: translation rate | ||
| + | Functions | ||
| + | *We model the production function of mRNA as a Hill-like, sigmoidal function | ||
[https://2008.igem.org/Team:Michigan/Project/Modeling Back to Modeling] | [https://2008.igem.org/Team:Michigan/Project/Modeling Back to Modeling] | ||
|} | |} | ||
Revision as of 02:16, 30 October 2008
|
|---|
|
Sequestillator Model 2: A More Complicated ModelWhile Model 1 gave us some important results (mainly: need a low Kd for oscillations to occur) , we decided to look at more complete model that accounted for some of the dimerizations the proteins undergo: Parameters
Functions
|
|---|
 "
"