Team:NTU-Singapore/Modeling
From 2008.igem.org
Lalala8585 (Talk | contribs) |
Lalala8585 (Talk | contribs) |
||
| Line 145: | Line 145: | ||
|} | |} | ||
| - | + | ==E7 prodcution system== | |
| + | {| align="center" border="1" | ||
| + | |Type | ||
| + | |Parameter | ||
| + | |Values | ||
| + | |Comments | ||
| + | |- | ||
| + | |Phosphorylation Rate of Ai-2 | ||
| + | |kPF | ||
| + | |1 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Dephosphorylation Rate of Ai-2 | ||
| + | |kPB | ||
| + | |1 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Transcription Rate of LsrR gene | ||
| + | |k1C | ||
| + | |4.402517 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Transcription Rate of SupD gene | ||
| + | |k1D | ||
| + | |46.667 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Transcription Rate of t7pTag gene | ||
| + | |k1E | ||
| + | |1.5556 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Transcription Rate of Lysis gene | ||
| + | |k1F | ||
| + | |28 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Degradation Rate of LsrR mRNA | ||
| + | |d1C | ||
| + | |0.159845 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Degradation Rate of SupD mRNA | ||
| + | |d1D | ||
| + | |1.694359 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Degradation Rate of t7pTag mRNA | ||
| + | |d1E | ||
| + | |0.056478 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Degradation Rate of Lysis mRNA | ||
| + | |d1F | ||
| + | |1.0166 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Translation Rate of LsrR Protein | ||
| + | |k2C | ||
| + | |7.54716 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Translation Rate of t7 pTag Protein | ||
| + | |k2E | ||
| + | |2.6667 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Translation Rate of Lysis Protein | ||
| + | |k2F | ||
| + | |48 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Protein Degradation Rate | ||
| + | |d2C,d2E,d2F | ||
| + | |0.03465 | ||
| + | |Made using Earlier assumptions | ||
| + | |- | ||
| + | |Hill coefficeint for SupD | ||
| + | |nD | ||
| + | |50 | ||
| + | |Trial And Error | ||
| + | |- | ||
| + | |Hill coefficeint for t7 pTag | ||
| + | |nE | ||
| + | |1 | ||
| + | |Trial And Error | ||
| + | |- | ||
| + | |Hill coefficeint for Lysis | ||
| + | |nF | ||
| + | |1 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Dissociation Constant for SupD | ||
| + | |KD | ||
| + | |15 | ||
| + | |[5] | ||
| + | |- | ||
| + | |Dissociation Constant for t7 pTag | ||
| + | |KE | ||
| + | |1 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Dissociation Constant for Lysis | ||
| + | |KF | ||
| + | |0.8 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Constitutive Portion for SupD | ||
| + | |aD | ||
| + | |0.01 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Constitutive Portion for t7 pTag | ||
| + | |aE | ||
| + | |0.01 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Constitutive Portion for Lysis | ||
| + | |aF | ||
| + | |0.0001 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Complex Formation Rate Between <br> LsrR Repressor and Ai-2-Phosphorylated | ||
| + | |k3BC | ||
| + | |0.01 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Complex Formation Rate Between <br> SupD tRNA and t7 pTag mRNA | ||
| + | |k3DE | ||
| + | |0.000000001 | ||
| + | |Estimate | ||
| + | |- | ||
| + | |Amount of Protein Kinase | ||
| + | |S | ||
| + | |28.747 | ||
| + | |From Earlier Assumptions | ||
| + | |- | ||
| + | |} | ||
| + | |||
=Results of Modeling= | =Results of Modeling= | ||
Revision as of 02:42, 25 June 2008
|
Contents |
Introduction
The use of models to describe synthetic biology has its merits. Synthetic biology investigates the use of different biological parts to put together and assemble devices that carry out specific functions. Good mathematical models to describe each part would greatly help not only in the characterization of a part but also facilitate the use of the part by other people when they choose to use the part within their devices or systems.
Simulations based on modeling can give a first insight on how the system would turn out and provide a rough guide of the system’s behaviour.
System
The system can be viewed as two parts. The first part comprises of lactose induced production of colicin E7 and the immunity protein. The second part comprises of a detection mechanism that produces the lysis protein upon the detection of both Iron ions and Ai-2 ( Autoinducer 2).
ODEs used in modeling
The following equations shows the break down of the different equations that will be used in this modeling exercise. By understanding this section, it would make the understanding of the system of ODEs used
Constant synthesis & Linear Synthesis
- Simple ode to describe constant synthesis
- Gives an explicit analytical solution
- Unique solution once a IC is posed
Linear Degradation
- Rate of degradation is proportional to how much of the molecule is present
- Gives an explicit analytical solution
- Constant half life
Simple Forward Reaction
This equation ignores the fact that dissociation of the complex occurs. We can do so if the dissociation is much slower than the formation.
- Single solvable equation for the unknown C
- Simple, unique solution available with I.C
Phosphorylation and Dephosphorylation
Assumptions:
- Linear kintic rate laws apply only if XT is much less than the Michaelis constants of both kinase and phosphotase
- Modeled after simple linear kinetics
- Gives a hyperbolic signal response curve when X plotted vs S
Regulated Transcription
ODE system used in model
Lactose controlled production of E7 + Imm
Variables
- LacI = A
- Lactose =B
- E7 = C
Iron and Ai2 controlled production of Lysis
Variables
Variables
- Ai-2 : A
- ai-2-phos : B
- LsrR : C
- SupD derivatives : D
- T7ptag : E
- Lysis : F
Parameter Estimation
Estimation of different parameters
- Transcription : 70nt/s
- Translation : 40aa/s
- Number of Essential Genes : 297
- Number of mRNA per cell : 4000
- Average mRNA half life : 5min
- Average mRNA length : 1100
Assumptions
- Rate of transcription is dependent on length of gene
- Number of amino acids is 1/3 of the number of nucleotides in a gene
- Rate of Translation is dependent on number of nucleotides
- For each gene mRNA = 10 at steady state
- Rate of degradation of average mRNA = 1100/ 5 min
- Rate of degradation of protein is equivalent to time for cell division i.e. 40 min
E7 prodcution system
| Type | Parameter | Values | Comments |
| Transcription Rate of Lac I gene | k1A | 21 | Made using Earlier assumptions |
| Transcription Rate of E7 + Imm gene | k1C | 2.470588 | Made using Earlier assumptions |
| Degradation Rate of Lac I mRNA | d1A | 0.76246 | Made using Earlier assumptions |
| Transcription Rate of E7 + Imm mRNA | d1C | 0.0897 | Made using Earlier assumptions |
| Translation Rate of Lac I mRNA | k2A | 36 | Made using Earlier assumptions |
| Translation Rate of E7 + Imm mRNA | k2C | 4.23539 | Made using Earlier assumptions |
| Protein Degradation Rate | d2A,d2C | 0.03465 | Made using Earlier assumptions |
| Hill coefficeint for E7 + Imm | nC | 1 | This is obtained on the assumption that one Repressor Protein binds to one Lactose molecule complex |
| Dissociation Constant for E7 + Imm | KC | 0.8 | [3] |
| Constitutive Portion for E7 + Imm | aC | 0.5 | Estimate since a is between 0 and 1 Implication that Lactose may not be a very strongly Regulated Promoter |
| Complex Formation Rate Between Lac I Repressor and Lactose | k3AB | 1 | Estimate |
E7 prodcution system
| Type | Parameter | Values | Comments |
| Phosphorylation Rate of Ai-2 | kPF | 1 | Estimate |
| Dephosphorylation Rate of Ai-2 | kPB | 1 | Estimate |
| Transcription Rate of LsrR gene | k1C | 4.402517 | Made using Earlier assumptions |
| Transcription Rate of SupD gene | k1D | 46.667 | Made using Earlier assumptions |
| Transcription Rate of t7pTag gene | k1E | 1.5556 | Made using Earlier assumptions |
| Transcription Rate of Lysis gene | k1F | 28 | Made using Earlier assumptions |
| Degradation Rate of LsrR mRNA | d1C | 0.159845 | Made using Earlier assumptions |
| Degradation Rate of SupD mRNA | d1D | 1.694359 | Made using Earlier assumptions |
| Degradation Rate of t7pTag mRNA | d1E | 0.056478 | Made using Earlier assumptions |
| Degradation Rate of Lysis mRNA | d1F | 1.0166 | Made using Earlier assumptions |
| Translation Rate of LsrR Protein | k2C | 7.54716 | Made using Earlier assumptions |
| Translation Rate of t7 pTag Protein | k2E | 2.6667 | Made using Earlier assumptions |
| Translation Rate of Lysis Protein | k2F | 48 | Made using Earlier assumptions |
| Protein Degradation Rate | d2C,d2E,d2F | 0.03465 | Made using Earlier assumptions |
| Hill coefficeint for SupD | nD | 50 | Trial And Error |
| Hill coefficeint for t7 pTag | nE | 1 | Trial And Error |
| Hill coefficeint for Lysis | nF | 1 | Estimate |
| Dissociation Constant for SupD | KD | 15 | [5] |
| Dissociation Constant for t7 pTag | KE | 1 | Estimate |
| Dissociation Constant for Lysis | KF | 0.8 | Estimate |
| Constitutive Portion for SupD | aD | 0.01 | Estimate |
| Constitutive Portion for t7 pTag | aE | 0.01 | Estimate |
| Constitutive Portion for Lysis | aF | 0.0001 | Estimate |
| Complex Formation Rate Between LsrR Repressor and Ai-2-Phosphorylated | k3BC | 0.01 | Estimate |
| Complex Formation Rate Between SupD tRNA and t7 pTag mRNA | k3DE | 0.000000001 | Estimate |
| Amount of Protein Kinase | S | 28.747 | From Earlier Assumptions |
Results of Modeling
E7 prodcution system
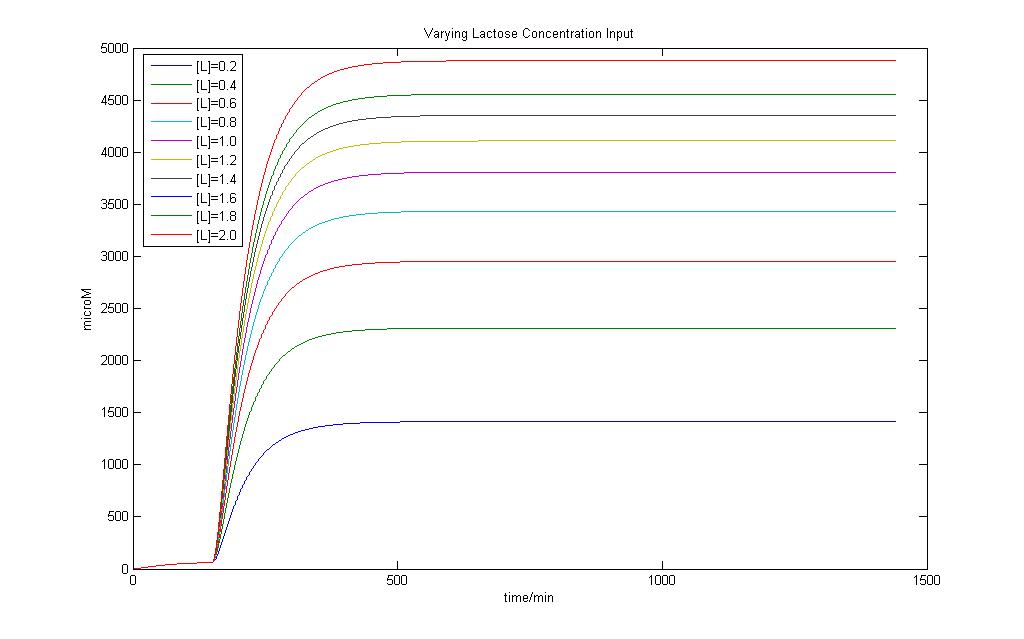
Modeling of the System shows that Lactose induction is essential to produce E7 and a variation of Lactose input can result in different yeilds of E7
Lysis Production system
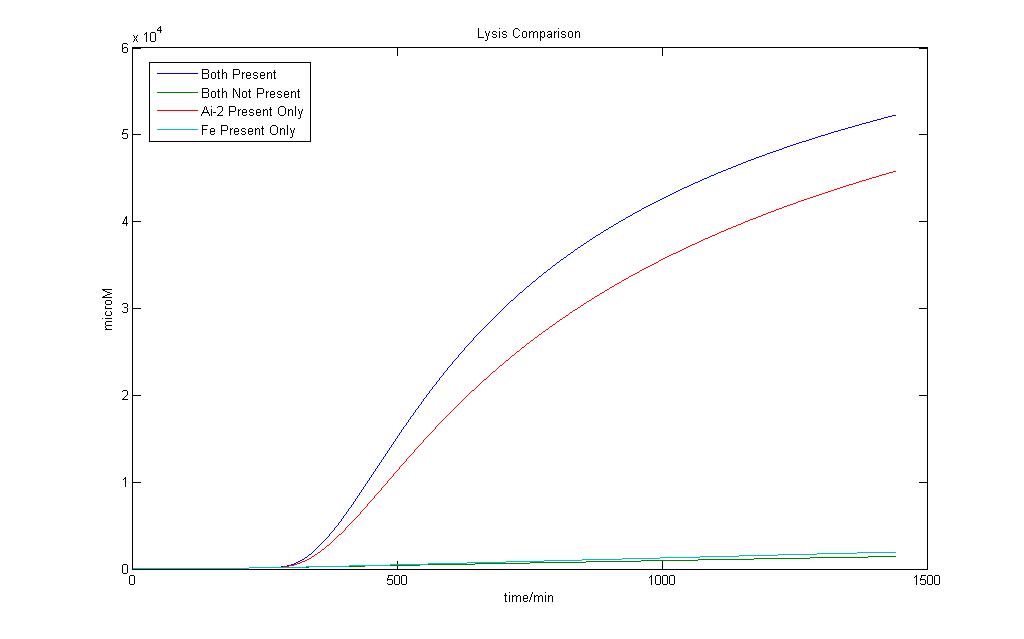
Here we define 1 as a certain threshold e.g. (<250 µM) that when the lysis protein reaches it, lysis in the cell definitely occurs. Although we wish for an absolute AND gate, where 0 will have no lysis production at all, simulation on biological systems shows that such results are impossible. Both addition Fe ions and Ai-2 alone would induce a certain level of lysis production. However when both are available, the lysis protein production would be higher.
In the presence of Ai-2 alone, Lysis will most likely still occur albeit at a slower rate compared to situation when both Fe and Ai-2 are present.
With the addition of iron and Ai-2 together the rate of lysis production is still significantly much higher compared to Ai-2 alone.
Therefore, by giving the logical output ‘1’ as a suitable threshold value of Lysis protein production higher than that of Ai-2 induction alone, we would still be able to obtain an AND gate based on our definition.
</div>
 "
"