Team:NTU-Singapore/Modelling/Stochastic Modeling
From 2008.igem.org
|
Contents |
Preliminary Run
| 
|
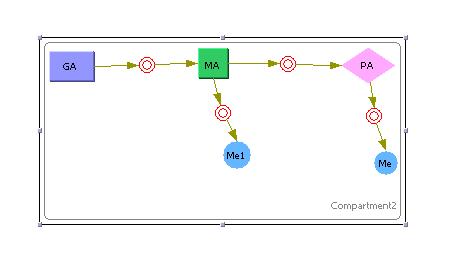
This model was generated using the program CellWare. The above model was simualted using a Hybrid ODE solver thats uses both deterministic and stochastic solvers. Work is now being carried out to see if this program can be used to build and simulate our systems.
All models below have been created using Cellware. Bionets2 is another option but we would first explore the use of the former program. The outputs are exported to MATLAB for plotting and visualisation.
Stochastic models using Gillispie Algorithm don't seem to work well as they seem to require a lot of computing power. Hybrid Stochastic ODEs seems to work better but the accuracy of the results seems to be sacrificed.
It is advised that the viewer observe the trends rather than the actual values given in the models. The modelin results would give us more of an idea of how the system would react rather than tell us exactly how much of a particular protein would be expressed. We hope that by doing this modeling exercise, we cna learn more about our system.
Stochastic Model For E7 + Imm Production
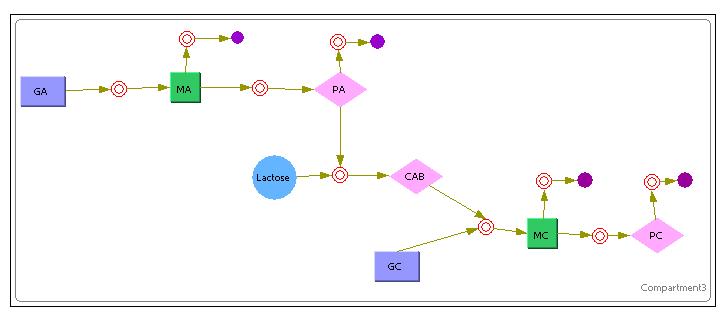
The above image shows the model that was used in Cellware for the 1st System.
The Purple Rectangles represent the DNA.
The Green Rectangles represent the mRNA.
The Pink Diamonds represent the Protein.
The Red Circles represent a certain reaction(Mass Action Reactions only since its a requirement for Stochastic simulations)
The Purple Full Circles represented degradation of a particular substance.
Unlike the previous Simulink deterministic models, Cellware does not allow the user to inject certain variables at stipulated times. Therefore, the model shows inoculation of Lactose at the VERY START of the simulation. This is equivalent to exposing a newly divided cell to an environment of lactose rather than allowing it time to reach a certain steady state for its constitutive proteins. Nevertheless, we hope that the model can still allow us some insights into the system.
| Parameter | Deterministic(m/l) | Stochastic(No. of Molecules) |
| GA | 28 | 28 |
| GC | 28 | 28 |
| Lactose | 10 | 10 |
Before we actually use the program to run any form of stochastic simulations, it would be prudent to check how its deterministic models would turn out. By varying the amount of Lactose input, a few graphs were obtained and they are as shown.
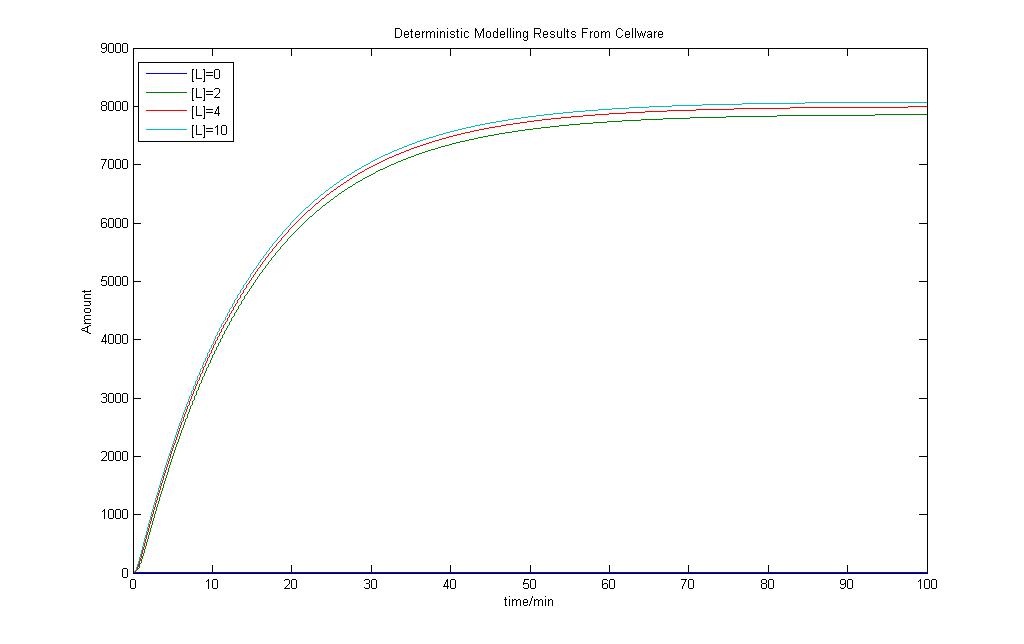
The model shows that there are differences in the output when the Lactose is changed but they are not exactly significant. However the model still shows very similar behaviour to the model we had obtained using Simulink earlier.
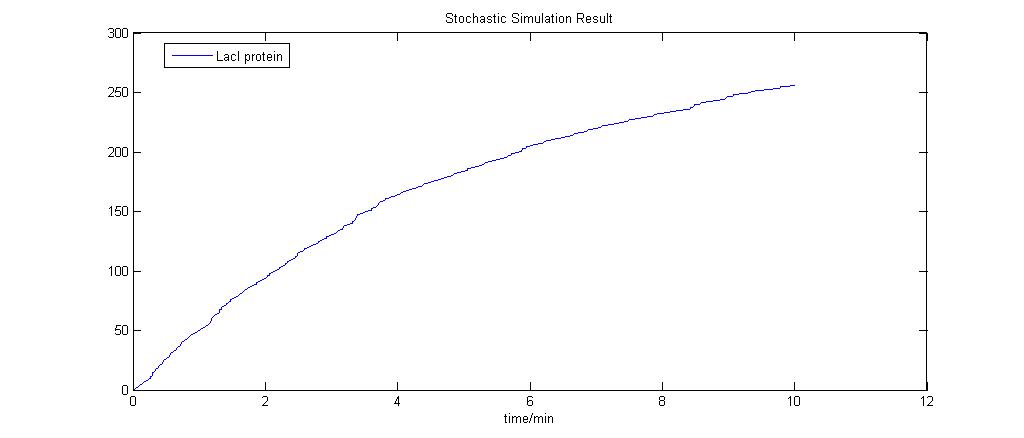
The stochastic modeling results are as shown and a total of 10 runs was done. The results are also compared to the Deterministic Modeling Output.
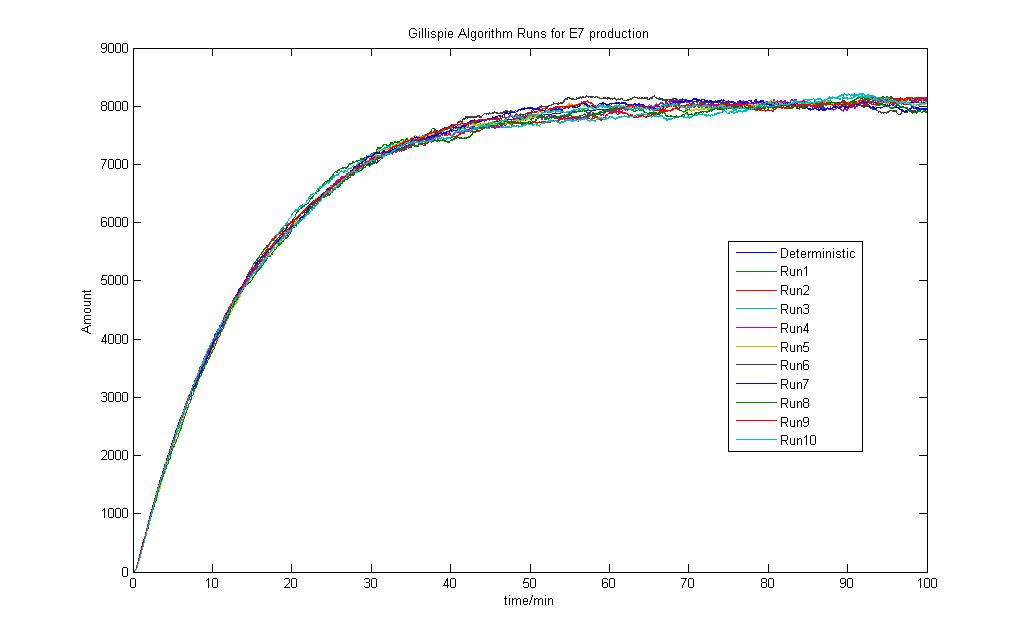
The run was done with 10mM lactose induction. As seen, the model is simple and the stochastic model produces results that are close to the deterministic model.
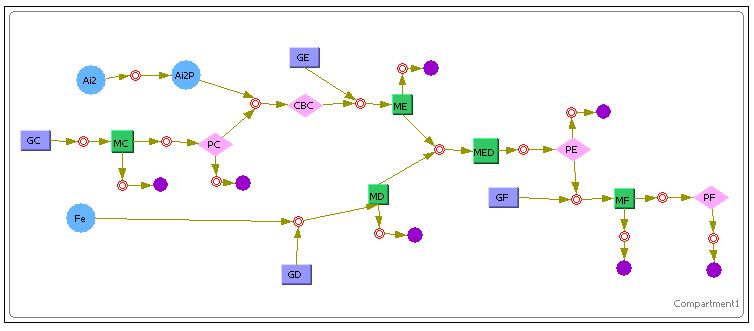
Stochastic Model For Lysis Production
The above diagram shows how we have modeled the Lysis Production system in Cellware. Again the legend is the same as the first E7 production system. We would also like to see how the Deterministic models turn out. The results are as shown.
| Parameter | Deterministic(m/l) | Stochastic(No. of Molecules) |
| GC | 85 | 10 |
| GD | 85 | 10 |
| GE | 85 | 10 |
| GF | 85 | 10 |
| Ai-2(Present) | 238 | 28 |
| Ai-2(Not Present) | 0 | 0 |
| Fe(Present) | 130.05 | 15.3 |
| Fe (Not Present) | 94.5 | 15 |
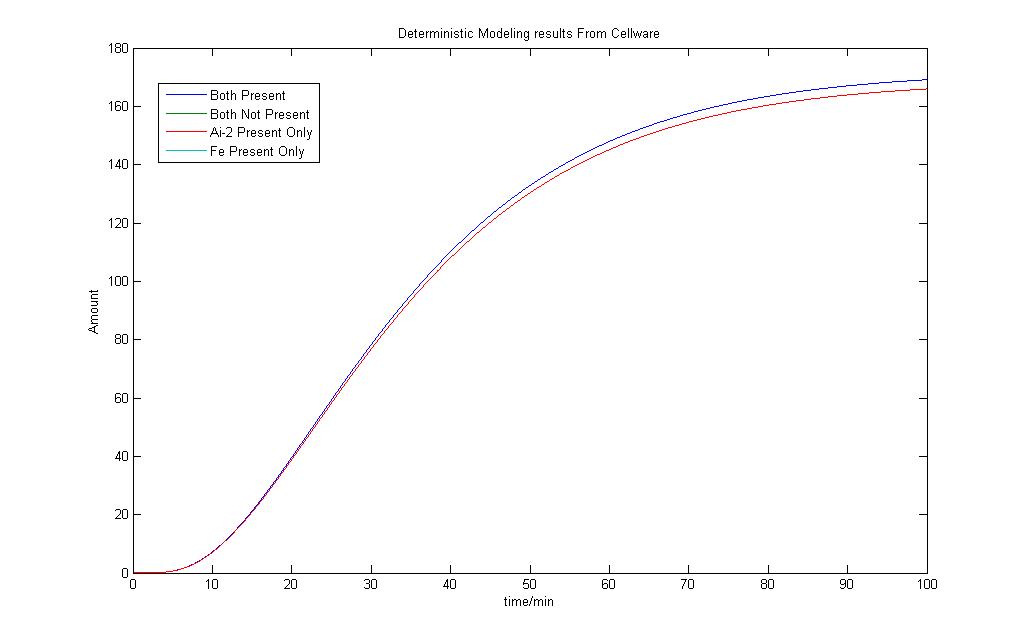
As we can see, a very similar outcome is observed as compared to the model we had obtained in Simulink. The difference in the Lysis protein output under the conditions that Ai-2 is present alone and when Iron and Ai-2 is present is not very pronounced.
For the Stochastic model, the number of molecules are also a guess and more work has to be done is refining the parameters. Presented here is the initial results for the condition that both Ai-2 and iron ions are present, thereby activating the and gate. The stochastic model results are also compared with the deterministic model.
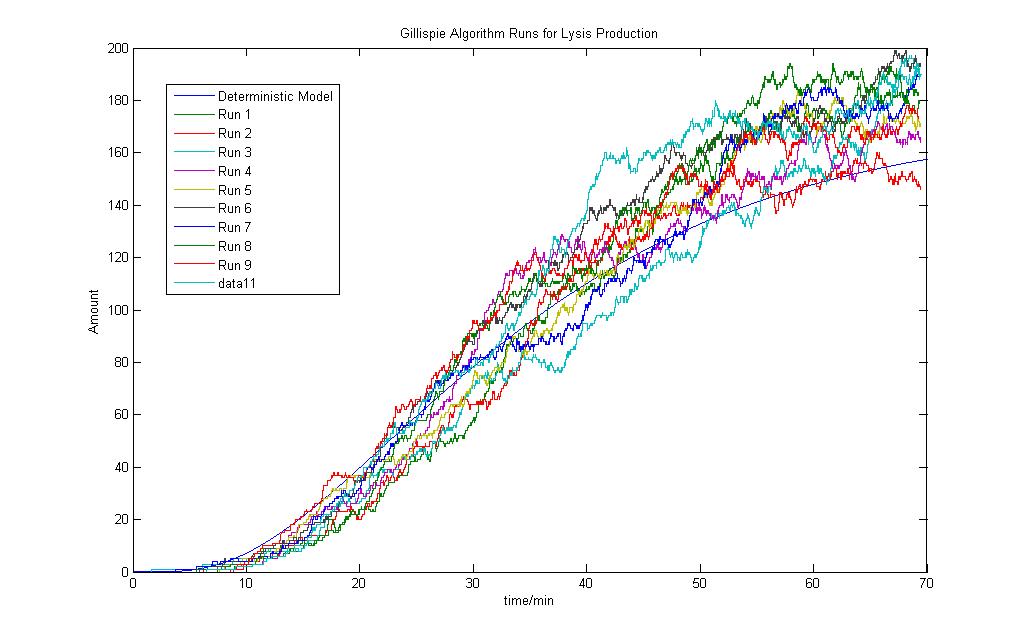
A total of 10 runs was done to observe the results. As expected, the random nature of the algorithm will give unique results for each run. Although the variations here are much larger compared to the first system, the general trend is still similar.
Updates
Currently the Simulations for the Lysis Production system tend to terminate prematurely at time = 70 min. The reason is still unknown but it could be due to the fact that the system is much more complicated than the E7 production system. Work is still being done to see if more models can be generated using Cellware. The program is generally easy to manipulate and the interface is quite user-friendly. If you have any comments on how our modeling exercise can be improved, don't hesitate to leave us an email . All contributions will be acknowledged by our team.
 "
"