Team:Newcastle University/Bug Busters
From 2008.igem.org
| Line 20: | Line 20: | ||
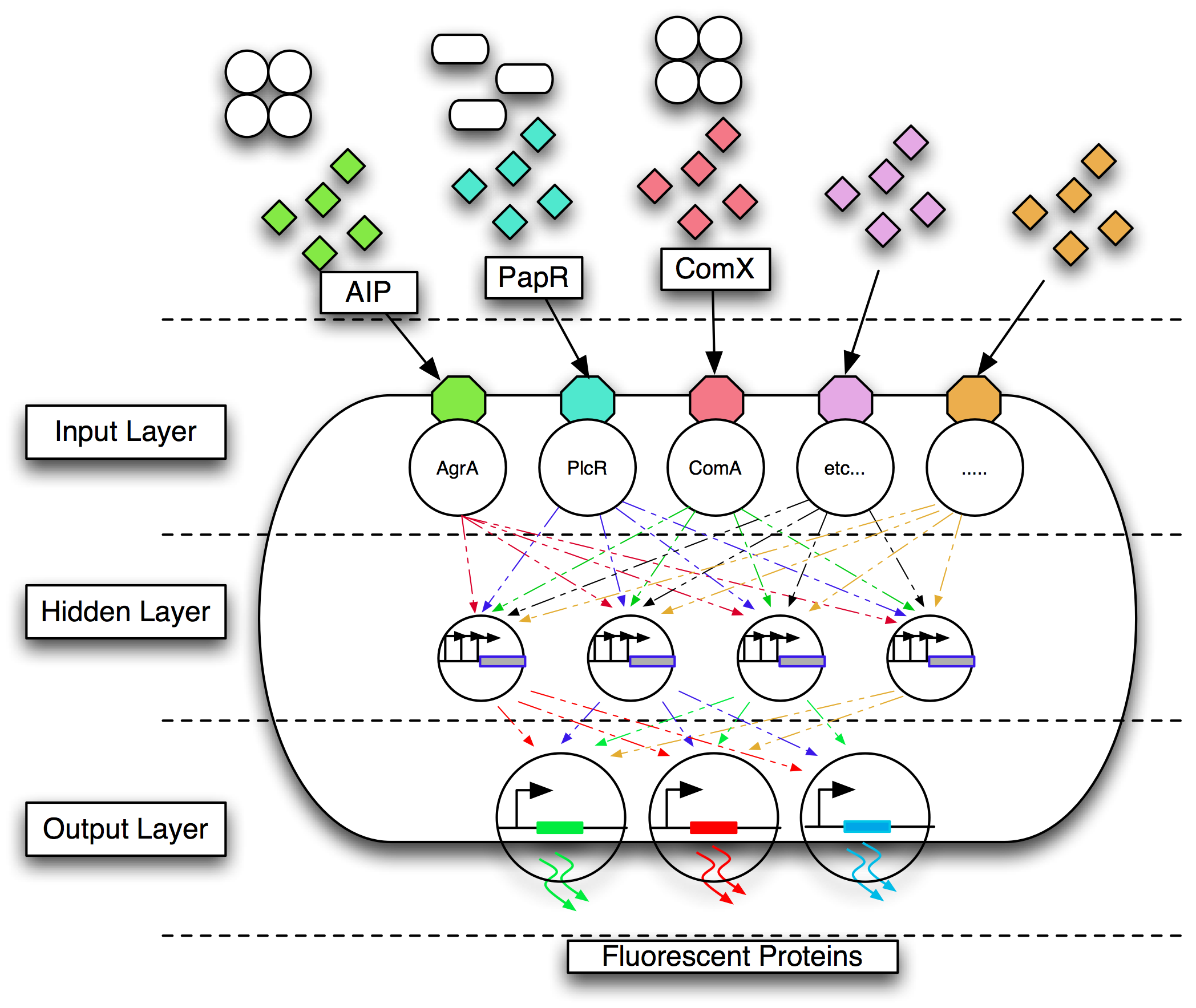
We are developing a diagnostic tool that enables the detection of a range of Gram-positive bacterial pathogens endemic in patients around the world. Currently, detection of these pathogens ranges from a few hours to several days. The system will use genetically engineered ''Bacillus subtilis'', to detect a range of Gram-positive bacterial pathogens. Our principal targets at this stage are ''Staphylococcus aureus'', and ''Streptococcus pneumoniae''. We are also considering the detection of a range of ''Bacillus'' species, and ''Clostridium difficile''. This new system will enable visual detection of these pathogens within minutes. The bacteria will be detected by the specific quorum-sensing peptides that they secrete extracellularly. Detection of the quorum-sensing peptides will activate the expression of fluorescent proteins, viewable under U.V. light, in our engineered ''B. subtilis'' chassis. This application would have importance not only in hospital settings, but also in the third world. | We are developing a diagnostic tool that enables the detection of a range of Gram-positive bacterial pathogens endemic in patients around the world. Currently, detection of these pathogens ranges from a few hours to several days. The system will use genetically engineered ''Bacillus subtilis'', to detect a range of Gram-positive bacterial pathogens. Our principal targets at this stage are ''Staphylococcus aureus'', and ''Streptococcus pneumoniae''. We are also considering the detection of a range of ''Bacillus'' species, and ''Clostridium difficile''. This new system will enable visual detection of these pathogens within minutes. The bacteria will be detected by the specific quorum-sensing peptides that they secrete extracellularly. Detection of the quorum-sensing peptides will activate the expression of fluorescent proteins, viewable under U.V. light, in our engineered ''B. subtilis'' chassis. This application would have importance not only in hospital settings, but also in the third world. | ||
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
</div> <!-- maincontent --> | </div> <!-- maincontent --> | ||
</html> | </html> | ||
Revision as of 14:46, 25 September 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Aim
Bug Busters
We are developing a diagnostic tool that enables the detection of a range of Gram-positive bacterial pathogens endemic in patients around the world. Currently, detection of these pathogens ranges from a few hours to several days. The system will use genetically engineered Bacillus subtilis, to detect a range of Gram-positive bacterial pathogens. Our principal targets at this stage are Staphylococcus aureus, and Streptococcus pneumoniae. We are also considering the detection of a range of Bacillus species, and Clostridium difficile. This new system will enable visual detection of these pathogens within minutes. The bacteria will be detected by the specific quorum-sensing peptides that they secrete extracellularly. Detection of the quorum-sensing peptides will activate the expression of fluorescent proteins, viewable under U.V. light, in our engineered B. subtilis chassis. This application would have importance not only in hospital settings, but also in the third world.
 "
"