Team:Newcastle University/Defining Parts
From 2008.igem.org
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Dry Lab >> Defining Parts
Introduction
Here we should start to suggest actual 'real' parts that we can use to implement our designed system. Its important we begin to define a list because:
- It will provide a list of parts for Megan to populate the repository
- A constrained parts list will narrow down the search space for the genetic algorithm
- It will provide a focus for the determining interactions and their parameters for the constraints repository.
- It will help push the design process
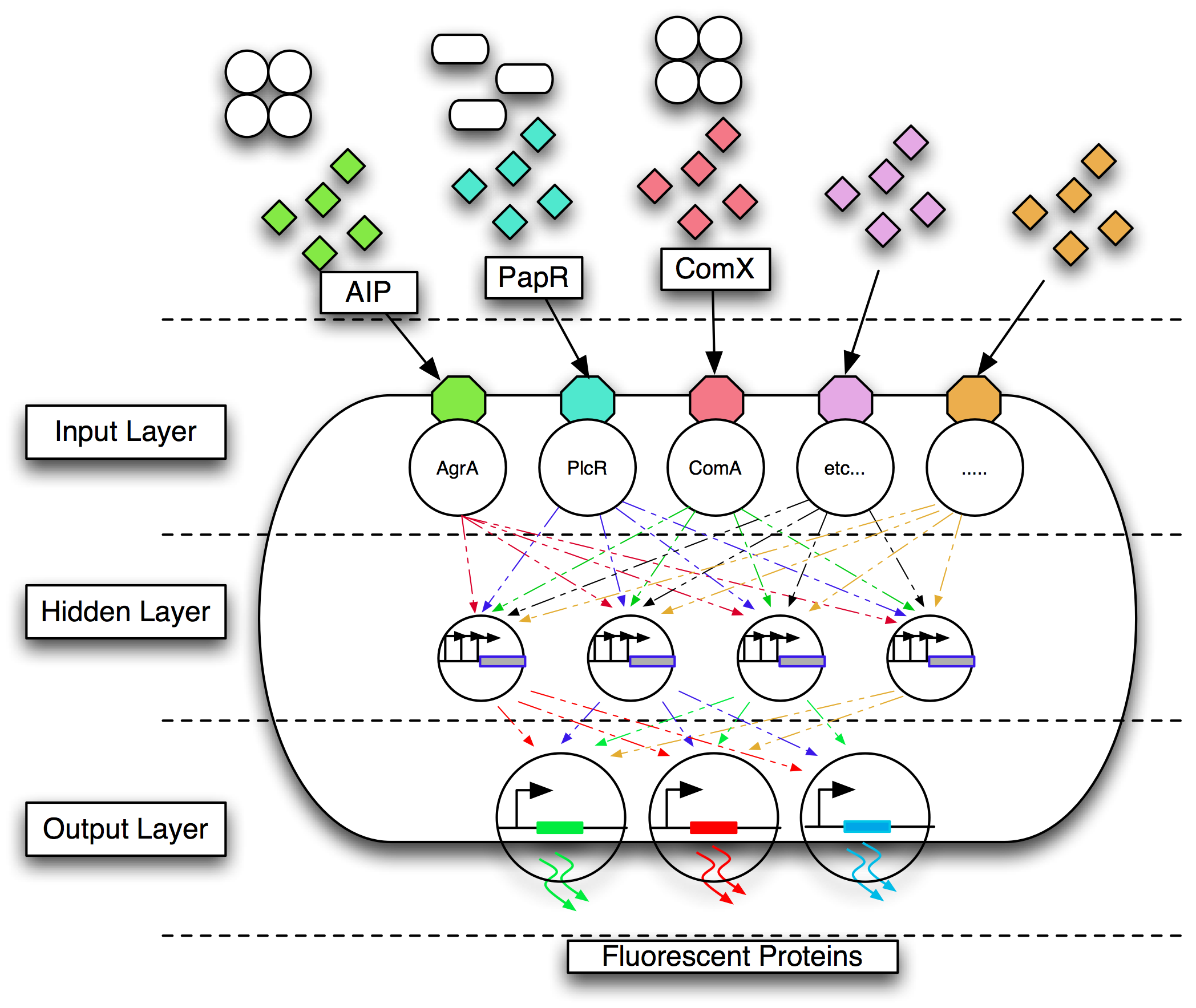
We intend to design and implement a neural network to classify the quorum sensing profile presented as an input. The neural network will produce a defined output. This involves parts for the input layer, hidden layer and the output layer.
It is important that the normal Bacillus signaling machinery doesn't interfere with the neural net and vice-versa. Therefore the components either need to be sourced from bugs sufficiently taxonomically different from Bacillus, from systems that Bacillus doesn't have or encapsulated. One idea is to use a sporulation deficient mutant. We can then use various sporulation and germination genes including sigma factors.
Jan-Willem has sent a list of possibilities for the parts list :
- pdr111_amye-phyper-spank_spec_.pdf (please contact Jan-Willem Veening)
- useful_bacillus_sequences.xls (please contact Jan-Willem Veening)
- Hans E. Plesser and Gaute T. Einevoll. Simulation of Biological Neural Networks. link
Input Layer
The parts for the input layer really depend on what we want to sense.
The way in which quorum sensing peptides are act as inputs to cellular signalling pathways depends on the actual peptide. Some such as the PapR peptide act directly to activate a transcription factor by binding to it. e.g. PapR binds to PlcR (see http://www.pnas.org/cgi/reprint/104/47/18490).
Others such as aip act by activating a two-component sensing system that in turn activates a transcriptional regulator e.g. See Cambridge 2007 entry.
We need an import system e.g. Opp to bring in the peptide. If this is non-specific we can possibly miss out the two component system.
Preliminary Parts List
| Part | Action | Target | Notes |
| LacI | repressor | LacO | Pspank |
| XylR | repressor | Pyxl | |
| LacO | repressor | ||
| Fi29 | |||
| Prophagepromoters | promoter | ||
| SSRA | tag | ||
| ComK | activator | PcomG | |
| RBS | |||
| Sigma factors | |||
| GFP | output | ||
| YFP | output | ||
| mCherry | output |
- Name = LacI
- UniqueID = x1234
- Type = Repressor protein
- Encoded by = y1234
- +Targets = LacO
- AaSequence = MKNQP
- Function = Repressor
- Model = (CellML)
- Name = LacI
- UniqueID = y1234
- Type = coding region
- Encodes = x1234
- Sequence = ACTGTGTG
- +Chassis =
- BioBrick/EMBL ID =
- Start and stop of BioBrick/EMBL ID =
- Function =
- Model = (CellML)
+ Possibly part of the constraints repository
 "
"