Team:Newcastle University/Notebook
From 2008.igem.org
Riachalder (Talk | contribs) |
Riachalder (Talk | contribs) |
||
| Line 340: | Line 340: | ||
Lane 1 = 1kb ladder | Lane 1 = 1kb ladder | ||
| + | |||
Lane 2 pGFP restricted with EcoRI and SpeI | Lane 2 pGFP restricted with EcoRI and SpeI | ||
| + | |||
Lane 3 = BBa_I746107 restricted with EcoRI and SpeI | Lane 3 = BBa_I746107 restricted with EcoRI and SpeI | ||
| - | This showed successful restriction of both plasmids which were then purified and ligated overnight using 6μl plasmid | + | This showed successful restriction of both plasmids which were then purified and ligated overnight using 6μl plasmid and 1.5μl enzyme (T4 ligase) in 25μl total reaction volume. |
</div><!--maincontent --> | </div><!--maincontent --> | ||
Revision as of 13:48, 10 September 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Wet Lab
Monday 4th August
We finally started in the labs today so we will start updating the wiki with what's been happening and the progress we are making.
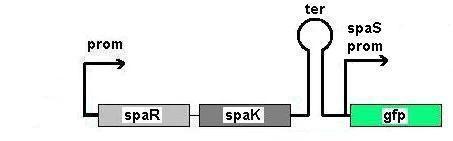
Fig 1: Newcastle iGEM team's construct
Fig 1 shows the construct which contains:
- spaRK promotor
- rrnB - rRNA binding site
- spaR (subtilin peptide antibiotic Regulation) - the 220 amino acid product of this gene usually regulates the downstream production of subtilin antibiotic. It has an N-terminal domain that can be phosphorylated and a C-terminal domian that has DNA binding properties [http://http://aem.asm.org/cgi/reprint/59/1/296.pdf]
- spaK (subtilin peptide antibiotic Kinase) - this gene codes for a 325 amino acid histadine kinase peptide that phosphorylates the N-terminus of spaR [http://http://aem.asm.org/cgi/reprint/59/1/296.pdf]. This activates the DNA binding ability of the C-terminus of spaR, which in turn initiates transcription of the downstream gene. In the case of our construct, this gene is gfp.
- gfp (green fluorescence protein) - the marker being used to show activation of the spaRK system and therefore diagnosis of gram-positive bacteria by B. Subtilis
- spaS promotor - a strong promotor inducible by upstream activation of spaRK. It can be placed in front any gene to regulate its activity.
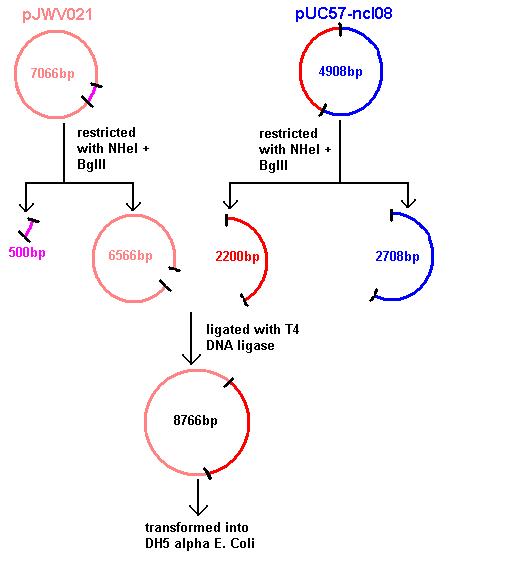
Aim 1: clone the spaRK system from pUC57 into pJWV021 and transform into DH5 alpha competent E. coli
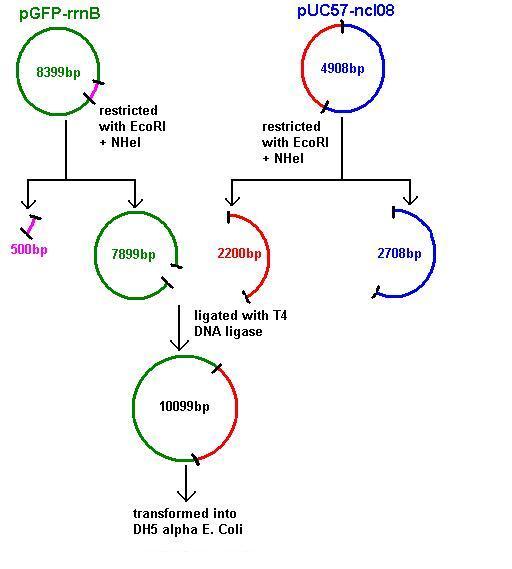
Aim 2: clone the spaRK system from pUC57 into pGFP-rrnB and transform into TOP10 competent E. coli
The 2.2kb fragment (ncl08) contains the spaRK system and promotor-less gfp linked to this. This means that when spaR is activated, its positive regulatory effect on spaK will in turn activate gfp.
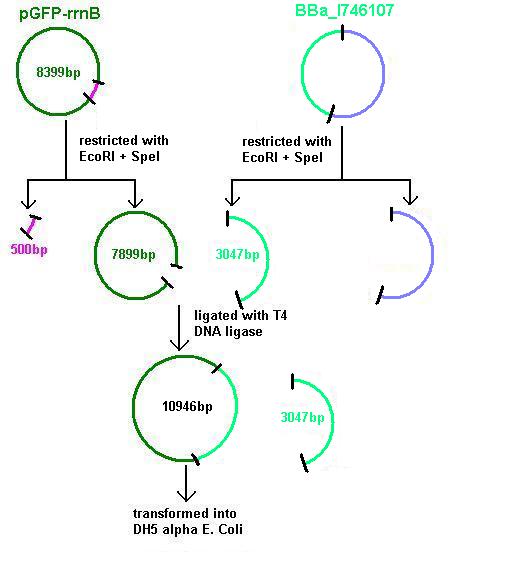
Aim 1: clone the agr system from the plasmid vector into pGFP-rrnB and transform into DH5 alpha competent E. coli
BBa_I647107 contains a partial agr operon, which includes agrC and A but has agrB and D deleted. This allows coding for the receptor (C/A) but not for production of the quorum peptides themselves (B/D). Eventually this will be linked to the spaRK system in a B. subtilis vector.
Monday 4th August
All 5 of us (Megan, Mark, Nina, Ria and Jess) went into the lab today to decide on a plan of action for the weeks to come.
- We used electrophoresis to check for the presence of plasmids (pGFPrrnB - our plasmid with the gfp gene, and pJWV021 - with the mCherry gene). Our gel was inconclusive, so we decided to re-run it the following day to confirm our results.
- In the afternoon we attempted to transform TOP10 competent E. coli cells with DNA from the registry (from source plate 1010 well 4A and 1018 7A).
- We followed the extraction method outlined in the green folder and for each DNA spot we plated two petri dishes: one with a larger volume of DNA (4μl) and one with a smaller volume (2μl) to make colony counting easier. We plated the following and incubated at 37˚C overnight:
Plate 1: +ve control (isolated plasmid plus TOP10 cells)
Plate 2: -ve control (TOP10 cells only, no plasmid)
Plate 3: Large 1010 4A
Plate 4: Small 1010 4A
Plate 5: Large 1018 7A
Plate 6: Small 1018 7A
Tuesday 5th August
Today we re-ran the gel electrophoresis of pGFP-rrnB and pJWV021 to check correct plasmid size and sufficient plasmid quantity. Results showed this to be true and the expected plasmid sizes of 8399bp for pGFP-rrnB and 7066bp for pJWV021.
Wednesday 6th August
Analyses of results from Monday’s transformation showed no colony growth except for the positive control. We decided to try another transformation into TOP10 E. coli using different spots from the registry. This time we used 1018 7A and 1004 6G.
We noticed that Cambridge, who had also been having problems, had adapted the extraction method. So we tried their improved method, with a couple of minor differences:
1) Warm 50μl of H20 in an Eppendorf tube at 50˚C
2) Add 4 punched-out spots from the registry
3) Keep at 50˚C for 20 minutes
4) Centrifuge at 13,300g for 3 minutes
5) Warm for a further 10 minutes at 50˚C
6) Centrifuge at 13,300g for 3 minutes
7) Pipette out the supernatant which should (hopefully!) contain the DNA
To see the original modified method, go to Cambridge’s OpenWetWare page:
http://openwetware.org/wiki/IGEM:Cambridge/2008/Protocols
Thursday 7th August
Unfortunately none of the plates showed any colonies. However, further incubation overnight of the plates at 37˚C from Monday 4th August produced 5 colonies on Plate 4 (2μl 1010 4A). These colonies were individually incubated overnight at 37˚C in 10μl LB.
Friday 8th August
The 5 colonies from Thursday 7th August cultured in LB were pooled and the plasmid purified. When the sample was run on gel no band appeared, showing that the colonies were not E. coli and must have resulted from contamination.
Monday 11th August
Plasmids were isolated from pUc57-ncl08 (the ncl08 fragment contains the spaRK 2-part component system). We aim to clone this into pGFP-rrnB and pJWV021 vectors.
Restrictions of pGFP-rrnB and pJWV021 carried out using 10μl plasmid in a total volume of 50μl. pJWV021 was restricted in a 2-step reaction as we beleived that the enzymes were incompatible in the same buffer. The second restriction of this plasmid used 48μl of the purified plasmid mixture in a total volume of 100μl. pGFP-rrnB was restricted using EcoRI and NHeI. pJWV021 was restricted firstly with NHeI and secondly with BglII.
These were purified and stored overnight at -25˚C.
Wednesday 27th August
Overnight cultures were analysed: - pGFP-rrnB grown on ampicillin - cells had lysed (medium was clear with large bubbles at the surface). - pJWV021 grown on ampicillin - frozen glycerol stock made (see protocols section). - pUC57-ncl08 grown on ampicillin - frozen glycerol stock made.
A colony stab was taken of pGFP-rrnB using 10μl spectinomycin in 10mL LB as the culture medium.
Thursday 28th August
New frozen glycerol stock of cells made to be kept at -80˚C.
Isolated plasmid from overnight culture of pGFP-rrnB. For this the 10mL culture was divided into 5 x 2mL plastic tubes and pelleted by centrifugation for 10 minutes. The supernatent was discarded and follwing the isolation procedure the product was pooled.
Friday 29th August
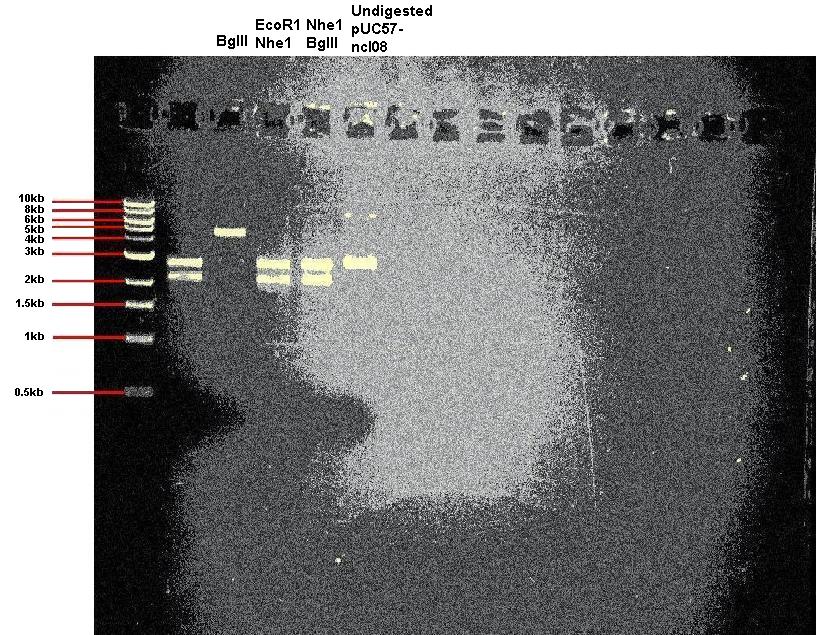
Today we digested our pUC57-ncl08 plasmid. In one reaction we used EcoR1 and Nhe1, and in other we used BglII and Nhe1. We also carried out a digestion with just BglII to check for the correct sized linear DNA. In addition we ran undigested plasmid on the gel.
Expected band sizes:
* For digestion with BglII only: 4.9kb
* For digestion with Nhe1 and EcoR1: 2.2kb and 2.7kb
* For digestion with Nhe1 and BglII: 2.2kb and 2.7kb
As you can see, the digests appear successful.
Thursday 04th September
The plates from 03.09.08 showed expected colony growth and therefore stab cultures were taken from individual colonies on Plates 1, 2 and 9. The culture media were made using 10mL of LB in 15mL plastic tubes together with the correct volume of the relavent antibiotic. This was 5μl spectinomycin for the GFP-rrnB colonies and 4μl kanomycin for the pJWV021 colonies. Cultures were taken as follows:
GFP-rrnB - pUC57 ligation (grown on spec)
- 9 white colonies (1-9) (Plate 1)
- 1 green colony (10) (Plate 1)
- 2 white colonies (11,12) (Plate 2)
pJWV021 - pUC57 ligation (grown on kan)
- 10 white colonies (13-22) (Plate 9)
- 1 pink colony (23) (Plate 9)
- 1 bright white colony (24) (Plate 9) (this is likely to a be contaminant non-E.coli colony)
All 24 culture tubes were incubated at 37˚C whilst shaking for ~20 hours. This large quantity of cultures should ensure that we obtain at least 2 cultures (one GFP-rrnB and one pJWV021) that have taken up the ncl08 insert. The plates were kept at room temperature in case fulrther stab cultures needed to be made.
Friday 05th September
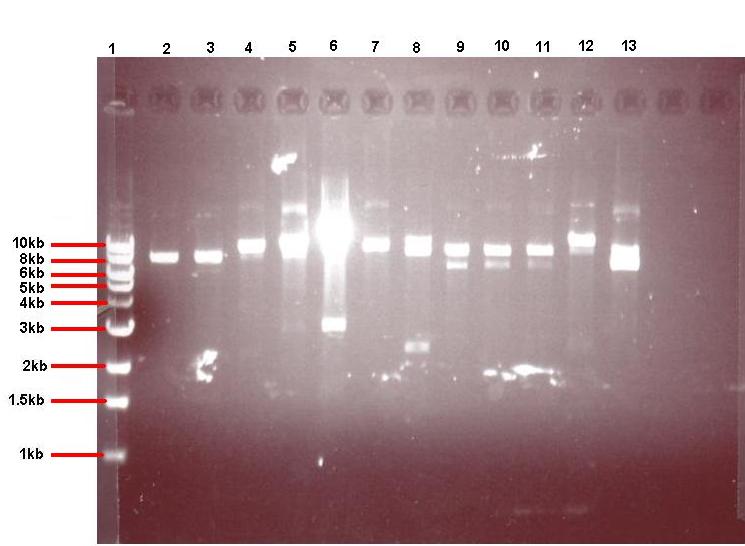
4μl of each of the 24 overnight cultures was centrifuged and the plasmid isolated from the resulting pellets. The isolated plasmid was then restricted to see if the plasmid had taken up the insert. All pGFPrrnB-nvl08 plasmids were restricted using EcoRI and NheI and pJWV021-ncl08 plasmids restricted with NheI and BglII. The restrictions were carried out in using 7.5μl plasmid in a total volume of 15μl. Samples were run on gel
Lane 1: 1kb ladder
Lane 2: pGFPrrnB-ncl08 colony 1 (white colony, 25μl plate)
Lane 3: pGFPrrnB-ncl08 colony 2 (white colony, 25μl plate)
Lane 4: pGFPrrnB-ncl08 colony 3 (white colony, 25μl plate)
Lane 5: pGFPrrnB-ncl08 colony 4 (white colony, 25μl plate)
Lane 6: pGFPrrnB-ncl08 colony 5 (white colony, 25μl plate)
Lane 7: pGFPrrnB-ncl08 colony 6 (white colony, 25μl plate)
Lane 8: pGFPrrnB-ncl08 colony 7 (white colony, 25μl plate)
Lane 9: pGFPrrnB-ncl08 colony 8 (white colony, 25μl plate)
Lane 10: pGFPrrnB-ncl08 colony 9 (white colony, 25μl plate)
Lane 11: pGFPrrnB-ncl08 colony 10 (white colony, 250μl plate)
Lane 12: pGFPrrnB-ncl08 colony 11 (white colony, 250μl plate)
Lane 13: pGFPrrnB-ncl08 colony 12 (green colony, 25μl plate)
Electrophoresis agarose gel showing restricted pGFPrrnB-ncl08 taken from 12 different colonies.
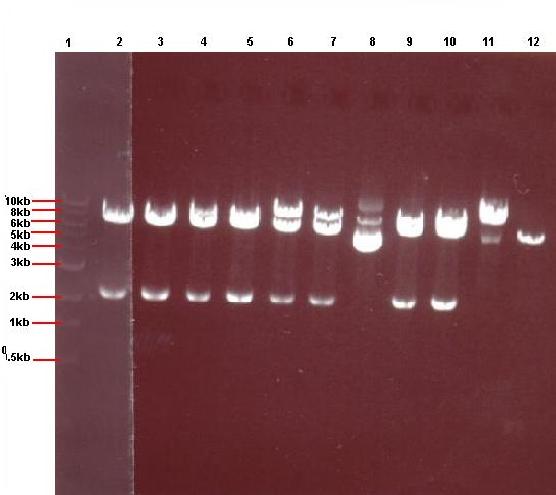
Lane 1: 1kb ladder
Lane 2: pJWV021-ncl08 colony 1 (white colony)
Lane 3: pJWV021-ncl08 colony 2 (white colony)
Lane 4: pJWV021-ncl08 colony 3 (white colony)
Lane 5: pJWV021-ncl08 colony 4 (white colony)
Lane 6: pJWV021-ncl08 colony 5 (white colony)
Lane 7: pJWV021-ncl08 colony 6 (white colony)
Lane 8: pJWV021-ncl08 colony 7 (white colony)
Lane 9: pJWV021-ncl08 colony 8 (white colony)
Lane 10: pJWV021-ncl08 colony 9 (white colony)
Lane 11: pJWV021-ncl08 colony 10 (white colony)
Lane 12: pJWV021-ncl08 colony 11 (white colony)
Lane 13: pJWV021-ncl08 colony 12 (white colony)
The gels showed correct insert fragments (2.2kb) in lanes 8 (colony 7) and 12 (colony 11) for pGFPrrnB and lanes 2, 3, 4, 5, 6, 7, 9 and 10 (colonies 13, 14, 15, 16, 17, 18, 20 and 21 respectively) for pJWV021.
Undigested plasmid from colonies 7 + 11 (pGFPrrnB) and 13 + 14 (pJWV021) were transformed into Bacillus subtilis (see protocol section).
Plasmid renamed once transformed into B. subtilis to avoid confusion with plasmid in E. coli:
- pGFPrrnB-ncl08 (E. coli) = iGEMgfp (B. subtilis) - pJWV021-ncl08 (E. coli) = iGEMcherry (B. subtilis)
Agar cultures were also made from the same colony cultures:
- iGEMgfp (chloramphenicol) colony 7
- iGEMgfp (chloramphenicol) colony 11
- iGEMgfp (chloramphenicol) negative control
- iGEMcherry (kanomycin) colony 13
- iGEMcherry (kanomycin) colony 14
- iGEMcherry (kanomycin) negative control
Monday 08th September
Results from the agar cultures:
- iGEMgfp (chloramphenicol) colony 7 - many colonies
- iGEMgfp (chloramphenicol) colony 11 - many colonies
- iGEMgfp (chloramphenicol) negative control - no colonies
- iGEMcherry (kanomycin) colony 13 - a few colonies
- iGEMcherry (kanomycin) colony 14 - 3 colonies (plus 'background' contaminants)
- iGEMcherry (kanomycin) negative control - no colonies (some 'background' contaminants)
Agar stabs were taken from the following colonies and streaked onto agar to obtain isolated colonies:
- iGEMgfp colonies 7.1, 7.2, 7.3, 7.4, 7.5, 7.6 - iGEMgfp colonies 11.1, 11.2, 11.3, 11.4, 11.5, 11.6 - iGEMcherry colonies 13.1, 13.2, 13.3 B. subtilis wild type strain ATCC6633 was also streaked onto agar.
Overnight (ON) cultures were made by JW of iGEMgfp 7.1, iGEMgfp colonies 11.1, iGEMcherry 13.1 and 13.2. The following cultures were then made and incubated at 37˚C whilst shaking for 3 hours. Dilute (dil) cultures were made by adding 500μl ON culture to 10mL LB. After 3 hours ATCC6633 supernatent was obtained by centrifuging ON ATCC6633 culture and 250μl of this added to the + ATCC6633 tubes.
- iGEMgfp ON + ATCC6633 - iGEMgfp ON - ATCC6633 - iGEMgfp dil + ATCC6633 - iGEMgfp dil - ATCC6633 - iGEMcherry ON + ATCC6633 - iGEMcherry ON - ATCC6633 - iGEMcherry dil + ATCC6633 - iGEMcherry dil - ATCC6633
These were left to incubate at 37˚C for a further 2 hours to allow gfp and mCherry induction by the subtilin contained int he ATCC6633 supernatent. Those wiothout supernatent were positive controls.
The B. subtilis cells were prepared for microscopy (see protocol section) and viewed:
- iGEMgfp ON + ATCC6633 - some cells expressed gfp - iGEMgfp ON - ATCC6633 - some cells expressed gfp (although less than in the presence of subtilin) - iGEMgfp dil + ATCC6633 - some cells expressed gfp - iGEMgfp dil - ATCC6633 - some cells expressed gfp (although less than in the presence of subtilin) - iGEMcherry ON + ATCC6633 - very few celled expressed mCherry - iGEMcherry ON - ATCC6633 - very few celled expressed mCherry - iGEMcherry dil + ATCC6633 - very few celled expressed mCherry - iGEMcherry dil - ATCC6633 - very few celled expressed mCherry
There appeared to be a difference in the GFP intensities between iGEMgfp with and without subtilin but this microspy was not sufficient to confirm this.
Starch, sucrose and glucose agar plates were made (see protocol section) and the following colonies streaked: - iGEMgfp 7.1, 7.2, 7.3, 7.4, 7.5, 7.6, 11.1, 11.2, 11.3, 11.4, 11.5, 11.6 streaked onto starch agar - iGEMcherry 13.1, 13.2, 13.3 streaked onto both glucose and sucrose
iGEMgfp should be able to grow on starch but will have no 'halo' surrounding the colonies (the halo is digested starch). This is becuase the insert should have integrated into amyE and the bacteria therefore cannot make amylase to digest starch. iGEMcherry should not be able to grow on sucrose as the insert should have integrated into sacA. The bacterium therefore cannot make sucrase to digest sucrose and consequently will have no carbon source. The glucose plates act as controls.
Overnigt cultures were also made og iGEMgfp 7.1 and 11.1 and iGEMcherry 13.1 and 13.2.
Tuesday 9th September
Half of the overnight cultures iGEMgfp 7.1 and iGEMcherry 13.1 were diluted (by adding 500μl ON culture to 10mL LB) to make the following cultures. These were incubated at 37˚C whilst shaking for 3 hours. 250μl ATCC6633 supernatent was then added to the + ATCC6633 tubes and the mixtures left to incubate for 2 hours at 37˚C.
Glycerol stocks made (see protocols section) of iGEMgfp 7.1 and 11.1 and iGEMcherry 13.1 and 13.2.
Microscopy of the cultures showed similar results to yesterday with again only a possible difference between the iGEMgfp with and without subtilin.
BBa_I746107 and pGFPrrnB restricted using 20μl plasmid and 2μl of each enzyme (EcoRI and SpeI) in 100μl total reaction volume. These were run on a gel at 70V for 1 hour.
Lane 1 = 1kb ladder
Lane 2 pGFP restricted with EcoRI and SpeI
Lane 3 = BBa_I746107 restricted with EcoRI and SpeI
This showed successful restriction of both plasmids which were then purified and ligated overnight using 6μl plasmid and 1.5μl enzyme (T4 ligase) in 25μl total reaction volume.
 "
"