Team:Newcastle University/Original Aims
From 2008.igem.org
MeganAylward (Talk | contribs) |
(represented vs represented by) |
||
| (26 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{:Team:Newcastle University/Header}} | |
| - | + | {{:Team:Newcastle University/Template:UnderTheHome|page-title=[[Team:Newcastle University/Original Aims|Original Aims]]}} | |
| - | + | == Original Aims == | |
| - | + | Bioinformatics is essential to modern biology, but is often considered as merely a tool, like a word processor or a spreadsheet, rather than an important approach to understanding and solving biological problems. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | We consider bioinformatics to be not only a tool in biology, but as a viable method for exploring a problem. Wet-lab biology is expensive in terms of time, money, and manpower. A single bioinformatician can investigate a given problem with no equipment other than a computer, and run many iterations of the same experiment within seconds, rather than the weeks that the labs may take. | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | However, the bioinformatics is only as good as its data and simulation power. The field expands daily with new information, all of which must be incorporated into a simulation in order for it to give useful results. Much of bioinformatics is backed by hard data from wet labs. Running an experiment 1,000 times is only useful if you know what all the variables are, and what they should be. | ||
| - | + | The most sensible approach to the biological investigations is to take advantahe both of of these different yet complementary methods - bioinformatics and wet lab experimentation - taking advantage of the strengths of each, while hopefully minimizing the limitations. | |
| - | == | + | ==BugBuster== |
| - | + | We aimed to develop a diagnostic biosensor for detecting pathogens. We wanted this to be cheaply and readily available for deployment in areas with limited medical resources such as refrigeration and sophisticated laboratories. We chose to use ''Bacillus subtilis'' as a method of delivery due to its ability to sporulate. The sensor bacteria could be dried down as spores, which are extremely resilient to environmental conditions, and could be rehydrated as required. | |
| - | + | ||
| - | + | Our aim was to develop a strain of [[Team:Newcastle University/Bacillus subtilis|''Bacillis subtilis 168'']] with the ability to detect four Gram-positive pathogens through their extracellularly-secreted quorum-sensing peptides, and to indicate through reporter genes linked to the receptor-ligand cascade which of these peptides are in its growth medium. | |
| + | |||
| + | Gram-positive bacteria communicate using quorum communication peptides. Research has shown that they are extremely strain-specific. ''B. subtilis'' has a range of genes that enable detection of these peptides. The issue that we had to overcome is the fact that we have more quorum-sensing peptides to detect than fluorescent proteins to show their presence. The output of the system is followed by analysing expression of fluorescent proteins such as mCherry, GFP, CFP and YFP. | ||
| + | |||
| + | Mapping multiple inputs to three output states is a multiplexing problem. The design of the genetic circuitry to do this is non-trivial and is not feasible manually. We therefore chose to use a biological implementation of an artificial neural network (ANN). Our team members wrote, designed and implemented a complete suite of tools that allowed the design and simulation of regulatory networks. An essential part of the approach was the use of computational evolution to design circuits with predictable behaviour even when the details of the required topology are unknown in advance. We used the modelling language CellML[http://www.cellml.org/] because of its ability to easily model virtual parts that can be easily assembled into a circuit which can be simulated. We aimed to translate this model into a sequence that could be implemented as one or more BioBricks. | ||
| + | |||
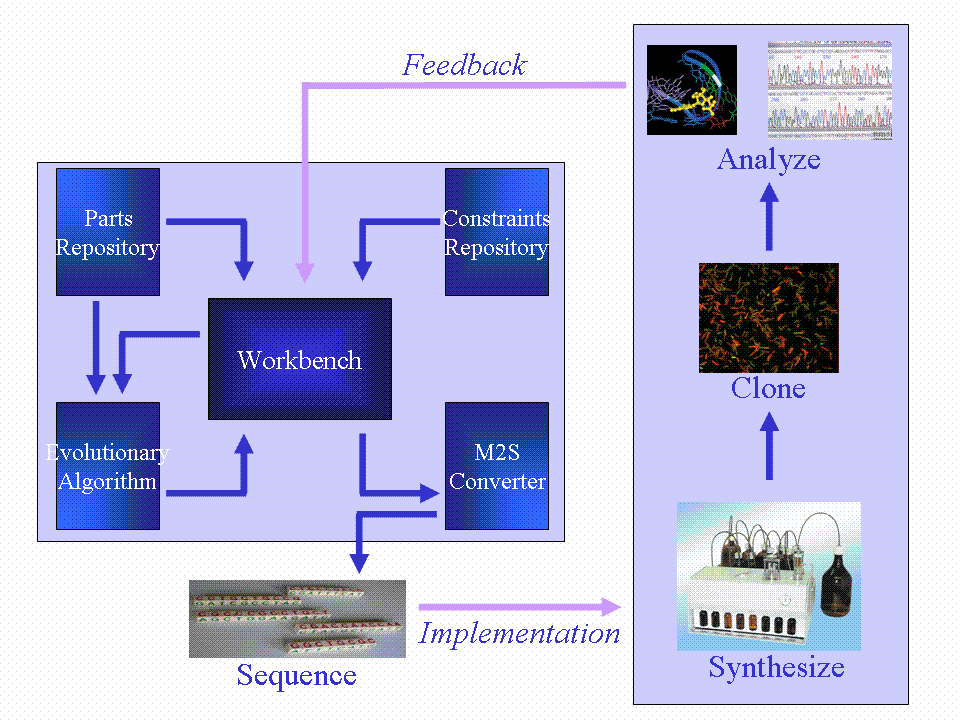
| + | [[Image:Overview.GIF|500px|center]] | ||
| + | |||
| + | <center>An overview of our complete system.</center> | ||
| + | |||
| + | A visual output was chosen for detection of pathogens, as this allows for rapid detection without a requirement for specialist equipment. The aim was to have different fluorescent protein outputs turned on by the presence of different pathogenic bacteria, and different combinations of these bacteria. In line with the neural network concept we wanted to map numerous inputs to limited outputs. Initially, we aimed to detect four pathogens, and we wanted different outputs for each of these and each combination of these. | ||
| + | |||
| + | We aimed to produce a [[Team:Newcastle University/Workbench|workbench]] that incorporates a [[Team:Newcastle University/Parts Repository|parts repository]], [[Team:Newcastle University/Constraints Repository|constraints repository]] and an [[Team:Newcastle University/Evolutionary Algorithm|evolutionary algorithm]] (EA). The EA takes input from the parts repository and constraints repository and evolves a neural network, using the CellML modelling language to carry out simulations of the network behaviour, which can be assessed for fitness (how closely the desired behaviour is reproduced). The fittest model can then be used to generate a DNA sequence implementing the neural network ''in vivo''. This DNA sequence can then be synthesized and cloned into the ''B. subtilis'' chassis. One of our outcomes should be a range of neural network node BioBrick devices which can be combined to form the ''in vivo'' neural network. | ||
| + | |||
| + | In keeping with the neural network model, our bacterial circuit would have three layers of complexity. The plan can be summed up as follows: | ||
| - | + | * The input layer to the bacterial neural network is represented by the two-component genes, activated by the peptides from the four gram-positive pathogens. Each peptide represents a node. | |
| + | * The hidden layer is represented by an assortment of different transcription factors. | ||
| + | * The output layer is represented by three fluorescent proteins; GFP, YFP and mCherry. | ||
| - | |||
| + | The biologist specifies the inputs and the outputs. In the presence of a particular profile of peptides, a specific fluorescent protein, or combination of proteins, will be expressed. | ||
| - | + | This was the starting point from which the construct was designed. | |
| - | + | ||
| - | + | ||
Latest revision as of 17:43, 29 October 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Original Aims
Original Aims
Bioinformatics is essential to modern biology, but is often considered as merely a tool, like a word processor or a spreadsheet, rather than an important approach to understanding and solving biological problems.
We consider bioinformatics to be not only a tool in biology, but as a viable method for exploring a problem. Wet-lab biology is expensive in terms of time, money, and manpower. A single bioinformatician can investigate a given problem with no equipment other than a computer, and run many iterations of the same experiment within seconds, rather than the weeks that the labs may take.
However, the bioinformatics is only as good as its data and simulation power. The field expands daily with new information, all of which must be incorporated into a simulation in order for it to give useful results. Much of bioinformatics is backed by hard data from wet labs. Running an experiment 1,000 times is only useful if you know what all the variables are, and what they should be.
The most sensible approach to the biological investigations is to take advantahe both of of these different yet complementary methods - bioinformatics and wet lab experimentation - taking advantage of the strengths of each, while hopefully minimizing the limitations.
BugBuster
We aimed to develop a diagnostic biosensor for detecting pathogens. We wanted this to be cheaply and readily available for deployment in areas with limited medical resources such as refrigeration and sophisticated laboratories. We chose to use Bacillus subtilis as a method of delivery due to its ability to sporulate. The sensor bacteria could be dried down as spores, which are extremely resilient to environmental conditions, and could be rehydrated as required.
Our aim was to develop a strain of Bacillis subtilis 168 with the ability to detect four Gram-positive pathogens through their extracellularly-secreted quorum-sensing peptides, and to indicate through reporter genes linked to the receptor-ligand cascade which of these peptides are in its growth medium.
Gram-positive bacteria communicate using quorum communication peptides. Research has shown that they are extremely strain-specific. B. subtilis has a range of genes that enable detection of these peptides. The issue that we had to overcome is the fact that we have more quorum-sensing peptides to detect than fluorescent proteins to show their presence. The output of the system is followed by analysing expression of fluorescent proteins such as mCherry, GFP, CFP and YFP.
Mapping multiple inputs to three output states is a multiplexing problem. The design of the genetic circuitry to do this is non-trivial and is not feasible manually. We therefore chose to use a biological implementation of an artificial neural network (ANN). Our team members wrote, designed and implemented a complete suite of tools that allowed the design and simulation of regulatory networks. An essential part of the approach was the use of computational evolution to design circuits with predictable behaviour even when the details of the required topology are unknown in advance. We used the modelling language CellML[1] because of its ability to easily model virtual parts that can be easily assembled into a circuit which can be simulated. We aimed to translate this model into a sequence that could be implemented as one or more BioBricks.
A visual output was chosen for detection of pathogens, as this allows for rapid detection without a requirement for specialist equipment. The aim was to have different fluorescent protein outputs turned on by the presence of different pathogenic bacteria, and different combinations of these bacteria. In line with the neural network concept we wanted to map numerous inputs to limited outputs. Initially, we aimed to detect four pathogens, and we wanted different outputs for each of these and each combination of these.
We aimed to produce a workbench that incorporates a parts repository, constraints repository and an evolutionary algorithm (EA). The EA takes input from the parts repository and constraints repository and evolves a neural network, using the CellML modelling language to carry out simulations of the network behaviour, which can be assessed for fitness (how closely the desired behaviour is reproduced). The fittest model can then be used to generate a DNA sequence implementing the neural network in vivo. This DNA sequence can then be synthesized and cloned into the B. subtilis chassis. One of our outcomes should be a range of neural network node BioBrick devices which can be combined to form the in vivo neural network.
In keeping with the neural network model, our bacterial circuit would have three layers of complexity. The plan can be summed up as follows:
- The input layer to the bacterial neural network is represented by the two-component genes, activated by the peptides from the four gram-positive pathogens. Each peptide represents a node.
- The hidden layer is represented by an assortment of different transcription factors.
- The output layer is represented by three fluorescent proteins; GFP, YFP and mCherry.
The biologist specifies the inputs and the outputs. In the presence of a particular profile of peptides, a specific fluorescent protein, or combination of proteins, will be expressed.
This was the starting point from which the construct was designed.
 "
"