Team:Paris/August 5
From 2008.igem.org
(Difference between revisions)
(→PCR Protocol) |
|||
| Line 1: | Line 1: | ||
| - | + | === Electrophoresis Purification of PCR=== | |
| - | + | ||
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
''When the PCR cycles were finished,'' | ''When the PCR cycles were finished,'' | ||
| Line 107: | Line 7: | ||
| - | After electrophoresis, the bands corresponding to | + | |
| + | After electrophoresis, the bands corresponding to the right amplification were excised and purified using the QIAquick DNA Gel Extraction Kit (QIAGEN). The elution was made in 50 µL of water. Because the intensity of the band corresponding to MP 120 was very low, we only continued with MP 100. MP 100 was digested by EcoRI & SpeI (Forward Insert) or by XbaI & PstI (D100 : Backward Insert). | ||
[[Image:KR000102.jpg|thumb|]] | [[Image:KR000102.jpg|thumb|]] | ||
| Line 113: | Line 14: | ||
[[Image:KR000106.jpg|thumb|]] | [[Image:KR000106.jpg|thumb|]] | ||
| - | === | + | {| |
| - | + | |align="center"|Name | |
| - | + | |align="center"|Promotor | |
| - | + | |align="center"|Gel | |
| - | + | |align="center"|Band | |
| - | + | |align="center"|Expected size | |
| - | + | |align="center"|Measured size | |
| - | + | |- | |
| - | + | |align="center"|PCR_124 | |
| - | + | |align="center"|pFlgA | |
| - | + | |align="center"| | |
| - | + | |align="center"| | |
| - | + | |align="center"| | |
| - | + | |align="center"| | |
| - | === | + | |- |
| - | + | |align="center"|PCR_125 | |
| - | + | |align="center"|pFlgB | |
| - | + | |align="center"| | |
| + | |align="center"| | ||
| + | |align="center"| | ||
| + | |align="center"| | ||
| + | |- | ||
| + | |align="center"|PCR_126 | ||
| + | |align="center"|pFlhB | ||
| + | |align="center"| | ||
| + | |align="center"| | ||
| + | |align="center"| | ||
| + | |align="center"| | ||
| + | |- | ||
| + | |align="center"|PCR_127 | ||
| + | |align="center"|pFlhDC | ||
| + | |align="center"| | ||
| + | |align="center"| | ||
| + | |align="center"| | ||
| + | |align="center"| | ||
| + | |} | ||
| - | + | ==> '''Remark :''' | |
Revision as of 14:01, 6 August 2008
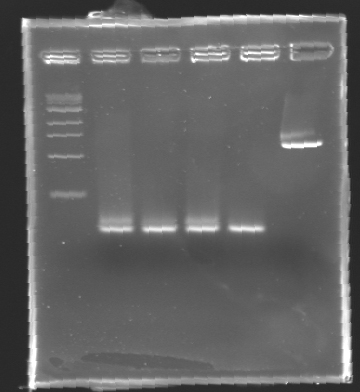
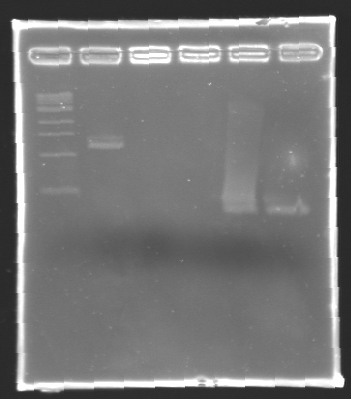
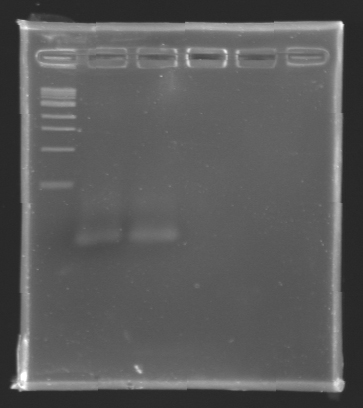
Electrophoresis Purification of PCR
When the PCR cycles were finished,
- 10 µL of 6X loading dye were added
- The samples were then loaded (2 x 30 µL per sample) on a 1,5% agarose gel.
After electrophoresis, the bands corresponding to the right amplification were excised and purified using the QIAquick DNA Gel Extraction Kit (QIAGEN). The elution was made in 50 µL of water. Because the intensity of the band corresponding to MP 120 was very low, we only continued with MP 100. MP 100 was digested by EcoRI & SpeI (Forward Insert) or by XbaI & PstI (D100 : Backward Insert).
| Name | Promotor | Gel | Band | Expected size | Measured size |
| PCR_124 | pFlgA | ||||
| PCR_125 | pFlgB | ||||
| PCR_126 | pFlhB | ||||
| PCR_127 | pFlhDC |
==> Remark :
 "
"