Team:Paris/Modeling/f3
From 2008.igem.org
(Difference between revisions)
| Line 42: | Line 42: | ||
|GFP concentration at steady-state | |GFP concentration at steady-state | ||
|nM | |nM | ||
| - | |need for 20 measures | + | |need for 20 + 20 measures <br> and 5x5 measures for the ''SUM''? |
|- | |- | ||
|(''fluorescence'') | |(''fluorescence'') | ||
| Line 63: | Line 63: | ||
|value | |value | ||
|- | |- | ||
| - | |β<sub> | + | |β<sub>12</sub> |
| - | |production rate of pFlhDC '''with RBS E0032''' <br> | + | |production rate of FliA-pFlhDC '''with RBS E0032''' <br> β<sub>12</sub> |
|nM.s<sup>-1</sup> | |nM.s<sup>-1</sup> | ||
| | | | ||
|- | |- | ||
| - | |(K<sub> | + | |(K<sub>11</sub>/{coef<sub>fliA</sub>}<sup>n<sub>11</sub></sup>) |
| - | |activation constant of | + | |activation constant of FliA-pFlhDC <br> K<sub>11</sub> |
| - | |nM<sup>n<sub> | + | |nM<sup>n<sub>11</sub></sup> |
| | | | ||
|- | |- | ||
| - | |n<sub> | + | |n<sub>11</sub> |
| - | |complexation order of | + | |complexation order of FliA-pFlhDC <br> n<sub>11</sub> |
|no dimension | |no dimension | ||
| | | | ||
|- | |- | ||
| - | |K<sub> | + | |- |
| - | | | + | |β<sub>2</sub> |
| - | |nM<sup>n<sub> | + | |production rate of OmpR-pFlhDC '''with RBS E0032''' <br> β<sub>2</sub> |
| + | |nM.s<sup>-1</sup> | ||
| + | | | ||
| + | |- | ||
| + | |(K<sub>11</sub>/{coef<sub>ompR</sub>}<sup>n<sub>19</sub></sup>) | ||
| + | |activation constant of OmpR-pFlhDC <br> K<sub>19</sub> | ||
| + | |nM<sup>n<sub>19</sub></sup> | ||
| | | | ||
|- | |- | ||
| - | |n<sub> | + | |n<sub>19</sub> |
| - | |complexation order | + | |complexation order of OmpR-pFlhDC <br> n<sub>19</sub> |
|no dimension | |no dimension | ||
| | | | ||
|} | |} | ||
Revision as of 14:45, 21 August 2008
At the steady-state, we haveand
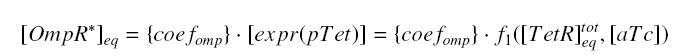
so the expression
gives
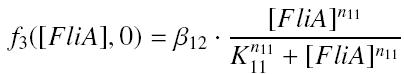
and
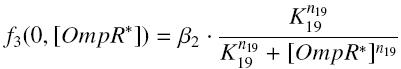
and for calculated values of the TF,

and
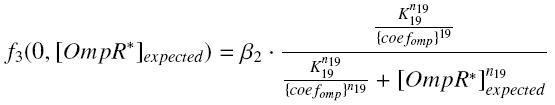
| param | signification | unit | value |
| [expr(pFlhDC)] | expression rate of pFlhDC with RBS E0032 | nM.s-1 | see "findparam" need for 20 + 20 measures and 5x5 measures for the SUM? |
| γGFP | dilution-degradation rate of GFP(mut3b) | s-1 | ln(2)/3600 |
| [GFP] | GFP concentration at steady-state | nM | need for 20 + 20 measures and 5x5 measures for the SUM? |
| (fluorescence) | value of the observed fluorescence | au | need for 20 + 20 measures and 5x5 measures for the SUM? |
| conversion | conversion ration between fluorescence and concentration | nM.au-1 | (1/79.429) |
| param | signification corresponding parameters in the equations | unit | value |
| β12 | production rate of FliA-pFlhDC with RBS E0032 β12 | nM.s-1 | |
| (K11/{coeffliA}n11) | activation constant of FliA-pFlhDC K11 | nMn11 | |
| n11 | complexation order of FliA-pFlhDC n11 | no dimension | |
| β2 | production rate of OmpR-pFlhDC with RBS E0032 β2 | nM.s-1 | |
| (K11/{coefompR}n19) | activation constant of OmpR-pFlhDC K19 | nMn19 | |
| n19 | complexation order of OmpR-pFlhDC n19 | no dimension |
 "
"