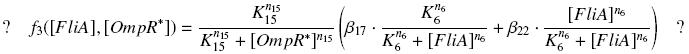Team:Paris/Modeling/f3
From 2008.igem.org
(Difference between revisions)
| Line 10: | Line 10: | ||
|unit | |unit | ||
|value | |value | ||
| + | |comments | ||
|- | |- | ||
|[expr(pFlhDC)] | |[expr(pFlhDC)] | ||
|expression rate of <br> pFlhDC '''with RBS E0032''' | |expression rate of <br> pFlhDC '''with RBS E0032''' | ||
|nM.min<sup>-1</sup> | |nM.min<sup>-1</sup> | ||
| - | | | + | | |
| + | |need for 20 mesures with well choosen values of [aTc]<sub>i</sub> <br> | ||
| + | and for 20 mesures with well choosen values of [aTc]<sub>i</sub> <br> | ||
| + | and 5x5 measures for the ''SUM''? | ||
|- | |- | ||
|γ<sub>GFP</sub> | |γ<sub>GFP</sub> | ||
| Line 20: | Line 24: | ||
|min<sup>-1</sup> | |min<sup>-1</sup> | ||
|0.0198 | |0.0198 | ||
| + | | | ||
|- | |- | ||
|[GFP] | |[GFP] | ||
|GFP concentration at steady-state | |GFP concentration at steady-state | ||
|nM | |nM | ||
| + | | | ||
|need for 20 + 20 measures <br> and 5x5 measures for the ''SUM''? | |need for 20 + 20 measures <br> and 5x5 measures for the ''SUM''? | ||
|- | |- | ||
| Line 29: | Line 35: | ||
|value of the observed fluorescence | |value of the observed fluorescence | ||
|au | |au | ||
| + | | | ||
|need for 20 + 20 measures <br> and 5x5 measures for the ''SUM''? | |need for 20 + 20 measures <br> and 5x5 measures for the ''SUM''? | ||
|- | |- | ||
| Line 35: | Line 42: | ||
|nM.au<sup>-1</sup> | |nM.au<sup>-1</sup> | ||
|(1/79.429) | |(1/79.429) | ||
| + | | | ||
|} | |} | ||
| Line 44: | Line 52: | ||
|unit | |unit | ||
|value | |value | ||
| + | |comments | ||
|- | |- | ||
|β<sub>12</sub> | |β<sub>12</sub> | ||
|production rate of FliA-pFlhDC '''with RBS E0032''' <br> β<sub>12</sub> | |production rate of FliA-pFlhDC '''with RBS E0032''' <br> β<sub>12</sub> | ||
|nM.min<sup>-1</sup> | |nM.min<sup>-1</sup> | ||
| + | | | ||
| | | | ||
|- | |- | ||
| Line 53: | Line 63: | ||
|activation constant of FliA-pFlhDC <br> K<sub>11</sub> | |activation constant of FliA-pFlhDC <br> K<sub>11</sub> | ||
|nM | |nM | ||
| + | | | ||
| | | | ||
|- | |- | ||
| Line 58: | Line 69: | ||
|complexation order of FliA-pFlhDC <br> n<sub>11</sub> | |complexation order of FliA-pFlhDC <br> n<sub>11</sub> | ||
|no dimension | |no dimension | ||
| + | | | ||
| | | | ||
|- | |- | ||
| Line 64: | Line 76: | ||
|production rate of OmpR-pFlhDC '''with RBS E0032''' <br> β<sub>2</sub> | |production rate of OmpR-pFlhDC '''with RBS E0032''' <br> β<sub>2</sub> | ||
|nM.min<sup>-1</sup> | |nM.min<sup>-1</sup> | ||
| + | | | ||
| | | | ||
|- | |- | ||
| Line 69: | Line 82: | ||
|activation constant of OmpR-pFlhDC <br> K<sub>19</sub> | |activation constant of OmpR-pFlhDC <br> K<sub>19</sub> | ||
|nM | |nM | ||
| + | | | ||
| | | | ||
|- | |- | ||
| Line 74: | Line 88: | ||
|complexation order of OmpR-pFlhDC <br> n<sub>19</sub> | |complexation order of OmpR-pFlhDC <br> n<sub>19</sub> | ||
|no dimension | |no dimension | ||
| + | | | ||
| | | | ||
|} | |} | ||
Revision as of 15:29, 9 October 2008
| param | signification | unit | value | comments |
| [expr(pFlhDC)] | expression rate of pFlhDC with RBS E0032 | nM.min-1 | need for 20 mesures with well choosen values of [aTc]i and for 20 mesures with well choosen values of [aTc]i | |
| γGFP | dilution-degradation rate of GFP(mut3b) | min-1 | 0.0198 | |
| [GFP] | GFP concentration at steady-state | nM | need for 20 + 20 measures and 5x5 measures for the SUM? | |
| (fluorescence) | value of the observed fluorescence | au | need for 20 + 20 measures and 5x5 measures for the SUM? | |
| conversion | conversion ratio between fluorescence and concentration | nM.au-1 | (1/79.429) |
| param | signification corresponding parameters in the equations | unit | value | comments |
| β12 | production rate of FliA-pFlhDC with RBS E0032 β12 | nM.min-1 | ||
| (K11/{coeffliA}) | activation constant of FliA-pFlhDC K11 | nM | ||
| n11 | complexation order of FliA-pFlhDC n11 | no dimension | ||
| β2 | production rate of OmpR-pFlhDC with RBS E0032 β2 | nM.min-1 | ||
| (K19/{coefompR}) | activation constant of OmpR-pFlhDC K19 | nM | ||
| n19 | complexation order of OmpR-pFlhDC n19 | no dimension |
Then, if we have time, we want to verify the expected relation
 "
"