Team:Paris/Perspectives
From 2008.igem.org
Philippe b (Talk | contribs) (→A.DNA nanocar factory) |
Philippe b (Talk | contribs) (→Biosynthetic Bottum-up approach) |
||
| Line 5: | Line 5: | ||
General comment: I am not at all against an explorative, speculative section. But it's purpose must be clear very clearly focussed on the interest of the FIFO for putative applications. Discuss each time why the FIFO could be especially interesting for your application. This must be very clear in order for this section to fulfill it's purpose.</font> | General comment: I am not at all against an explorative, speculative section. But it's purpose must be clear very clearly focussed on the interest of the FIFO for putative applications. Discuss each time why the FIFO could be especially interesting for your application. This must be very clear in order for this section to fulfill it's purpose.</font> | ||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 22:00, 29 October 2008
|
Un petit texte pour expliquer la motivation de cette page ne serait-ce que tout simplement: On this page we report some possible uses of the FIFO device in synthetic applications. General comment: I am not at all against an explorative, speculative section. But it's purpose must be clear very clearly focussed on the interest of the FIFO for putative applications. Discuss each time why the FIFO could be especially interesting for your application. This must be very clear in order for this section to fulfill it's purpose.
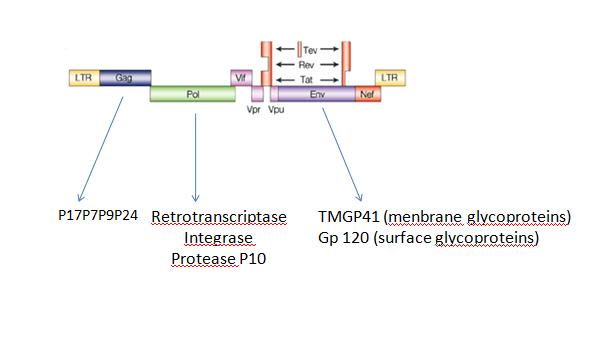
B.Artificial virus factoryWe can also produce a lot of differentes kinds of self-assembly structures like virus for example,HIV: No, you can not produce... You can say (possibly)... The FIFO device provides also a circuit useful to coordinate the self-assembly of molecular structure. A possible canevas for the production of synthetic virus? Indeed a virus such as HIV, displays ans organised sequential activation of genes that could be controled by a FIFO circuit. ARE YOU SURE OF THIS Fig2: genetic organization of HIV virus
In this setup I maybe understand the sequential order of activation, but I do not see why there should be the same order of inactivation. Justify please
If you ask us why? For 3 main reasons :
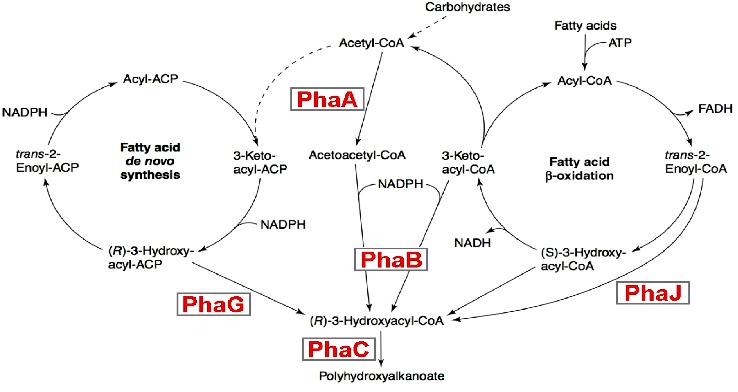
Metabolic engineering of polyhydroxyalkanoate biosynthesis pathwaysHuman overpopulation combined with the current lifestyle urges the rational, efficient, and sustainable use of natural resources to produce environmentally friendly plastic materials such as polyhydroxyalkanoic acids (PHAs), whose production/degradation cycle reduces undesirable wastes and emissions. Our study is a new metabolic strategy to generate PHA-hyperproducer strains, that have the properties to be a sequential metabolic pathway, we believe the sequential expression may increases the production of purified PHA. Our strategy consists on replacing the RFP,CFP and YFP genes by the PhaA ,PhaB and PhaC genes in our final system (containing oscillation,FIFO,synchronisation modules). This strategy is more efficient than a constitutive activation for 3 main reasons:
Again, you cannot just state this boldly without constructing an argument. You never explain why your system is better than simply expressing all the genes continuously at a lower level More generally, on this whole page: We don't expect you to give very detailed projects. You just need to give simple ideas for which the interest of the FIFO seems obvious (or at least you need to try making it seem obvious). We are not so much interested in the precise mechanisms or genes involved, but rather in the principles that make your projects interesting. Bibliography
Paul W. K. Rothemund
Paul W. K. Rothemund
Suvir Venkataraman, Robert M. Dirks, Paul W. K. Rothemund, Erik Winfree, Niles A. Pierce.
Georg Seelig, Bernard Yurke, Erik Winfree. |
 "
"