Team:Rice University/Notebook/18 July 2008
From 2008.igem.org
(Difference between revisions)
StevensonT (Talk | contribs) |
StevensonT (Talk | contribs) |
||
| Line 8: | Line 8: | ||
*Taylor Stevenson | *Taylor Stevenson | ||
*Repeat PCR from yesterday using these primers (NOTE: to view primers download this link and change extension to .doc (word file)[https://static.igem.org/mediawiki/2008/0/0d/Phage_round_1_primers.mov]). | *Repeat PCR from yesterday using these primers (NOTE: to view primers download this link and change extension to .doc (word file)[https://static.igem.org/mediawiki/2008/0/0d/Phage_round_1_primers.mov]). | ||
| - | * | + | **tried raising annealing temperature to 58*C |
[[Image:20080718.jpg|400px]] | [[Image:20080718.jpg|400px]] | ||
*#100bp standard | *#100bp standard | ||
Latest revision as of 21:13, 19 July 2008
Friday, July 18th
- David Ouyang
- Miniprep R0040 (ptet)
- Digest Amber Suppressor and ptet
- Ligate together
- Do ARFP mutant quikchange and DPN1
- Transform ARFP and Amber Suppressor supposed
- Taylor Stevenson
- Repeat PCR from yesterday using these primers (NOTE: to view primers download this link and change extension to .doc (word file)[1]).
- tried raising annealing temperature to 58*C
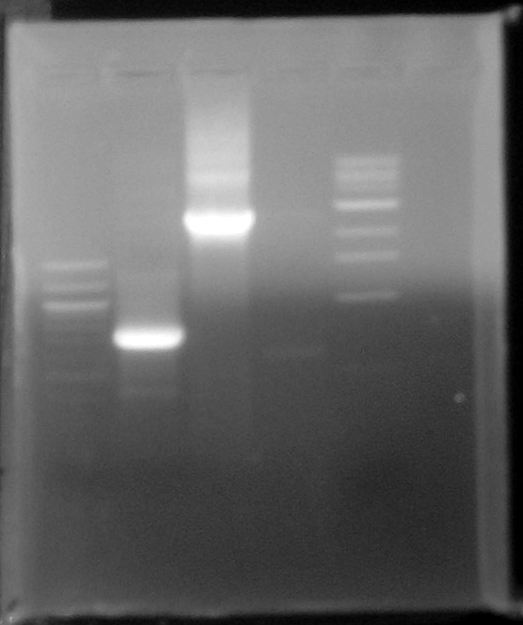
- 100bp standard
- BleoC PCR product
- Stf PCR product
- Ligation of yesterdays digested and CIPed PCR primers
- 1kb standard
- Result-The bands at 750bp and 2.5kbp correspond to the PCR products of BleoC_AvrII and Stf_NotI. These bands were cleaned, concentrated, and digested with NotI.
- The BleoC_AvrII digest was treated with CIP.
- Digested DNA was cleaned and concentrated, and ligated overnight at room temperature.
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
 "
"