Team:TUDelft/Temperature design
From 2008.igem.org
>> work in progress
Parts Design
First approach three excisting RNA thermometers and one artificial RNA thermometer based on G-quadruplex.
Scar problem
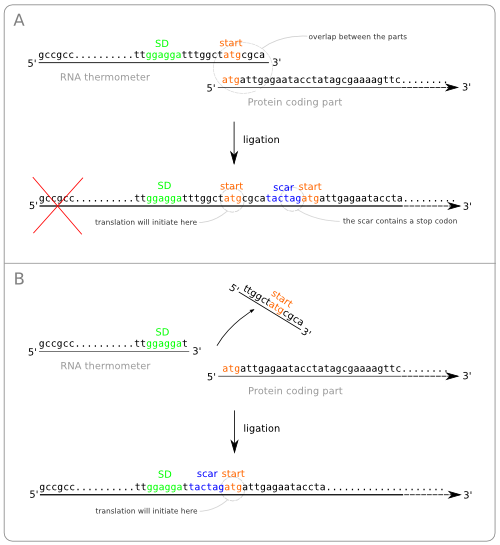
Turning the RNA thermometer sequences into standard biobricks is not as straightforward as adding a prefix and suffix to the found sequences. The reason for this is that the start codon of the protein coding part following the RNA thermometer is also part of the RNA thermometer sequence (figure ... sequences met atg overlap). The problem with this approach is that the ligation of the two parts results in a sequence containing two start codons with in between the scar (figure ... wrong situation sticking together two parts). The first of these start codon (the one on the RNA thermometer) will act as the initiation point for the translation, because of its distance to the Shine Dalgarno sequence. At first some extra amino acids will be added to the protein, but even worse, the protein will not be translated at all. This is because of the scar that contains a stop codon. So the translation will already end before the protein coding region is reached.
To solve this problem we have to alter the sequence of the RNA thermometer. Alteration of the protein coding part is of course no option because we are aiming for a standard part that can be combined with any other standardized protein coding part. So these parts, that always start with a start codon should be kept intact. The simple solution is to chop of the last part of the RNA thermometer sequence, which will be replaced by the scar and the start of the protein coding part after ligation (figure ... solution without scar adaption).
But this results in a new problem. The alteration of the RNA thermometer sequence also changes its secondary structure and that will also affect its function as temperature sensor (figure ... change of secondary structure). To resolve this we made some extra alteration in the RNA thermometer sequence at the opposite position of the scar in order to regain the original secondary structure.
We applied this procedure to three of the four RNA thermometers that we got from the literature. We used the Vienna RNAfold webserver to predict the secondary structure of the original RNA thermometer and the altered sequence. We regained the original secondary structure using a trial and error approach adding different nucleotides opposite of the added scar...
The fourth RNA thermometer, the artificial one based on the G-quadruplex structure, will loose its temperature sensitivity when the scar is introduced into the sequence. It happens to replace multiple G nucleotides that make up the G-quadruplex structure which is responsible for the temperature sensitivity.
Having designed sequences that have the same secondary structure does not guarantee the same functionality. The pairing nucleotides around the Shine Dalgarno region are changed, so the temperature needed to expose the Shine Dalgarno sequence will probably be changed. Tests will have to point out what the switching temperature of the RNA thermometer parts will be. This is of course if the parts work at all. There are probably a lot of factors (more than the predicted secondary structure can tell us) that play a role in the temperature sensing process, also many that we don't now of. Even one mutation can already lead ...
Changing the temperature threshold
Introduction...
In order to be able to design an RNA thermometer with a specific temperature threshold, we need to know which factors influence this threshold. The most important one is probably the secondary structure around the Shine Dalgarno sequence. Heat, or a rise in temperature, is needed to expose the Shine Dalgarno sequence by melting this part of the secondary structure. It thus sounds logical that the temperature threshold can be increased by incorporating nucleotides that form strong base pairs within the Shine Dalgarno region. This will result in a more stable helix that needs a higher temperature to become unstable, i.e. melt. Incorporating nucleotides that has weaker base-pairing strength within the Shine Dalgarno region will have the opposite effect. The helix will become less stable and less heat will be needed to melt it. This way the temperature threshold at which the translation will initiate will drop.
 "
"