Team:Tsinghua/Project
From 2008.igem.org
| HOME | Team | Project 1 | Project 2 | Parts | Modelling | Notebook | Doodle Board |
|---|
Contents |
Background
| Motile bacteria can respond to chemical gradients in their environment. Such directed movement in response to special chemicals is defined as chemotaxis. If organisms detect substances beneficial for surviving, positive chemotaxis occurs and cells will move in the direction of increasing concentration. In contrast, negative chemotaxis occurs when motile bacteria move away from a repellant. Both kinds of responses adjust the behaviors of organisms to the environment, thus improve their ability of surviving.
One typical chemotactic behavior is the response of Escherichia coli toward certain nutritions, which plays a key role in seeking food. Researchers have investigated thoroughly into the molecular mechanism of it.Motility in prokaryotes is accomplished by flagellum, a complex structure moving the cell by rotation, much like the propeller in a motor boat. In E.coli, counterclockwise rotation moves the cell in a direction called a run, while clockwise rotation results in tumbling of the cell.The direction of rotation is controlled by phosphorylation of CheY, a signaling protein in the chemotaxis pathway of E.coli. Low level of phosphorylation results in counterclockwise rotation of flagella and makes the cell move forward. In contrast, phosphorylated CheY favors tumbling. The level of protein phosphorylation is regulated by certain attractant or repellent, which can bind specific receptors on the cell surface. Signals are then passed through a series of intracellular biochemical reactions, in which phosphorylation of CheY is controlled. In this way bacteria accomplish chemotactic behavior. Escherichia coli is a common kind of engineer bacteria, which is convenient for genetic manipulation. It is likely that we make use of the chemotactic pathway in E.coli, making them detect and move toward a special chemical substance, maybe a certain kind of pollutant. If the genetic engineering strain also has the ability of degrading aimed chemicals, efficiency of pollution treatment can be hopefully improved a lot. However, the natural ligands concerning chemotaxis in E.coli are limited in several kinds of nutrition, such as carbohydrate and amino acids. Strains that can detecting more kinds of organic substances and even heavy metal ions is planed to be got through the reconstruction of chemotaxis system. |
Overall project
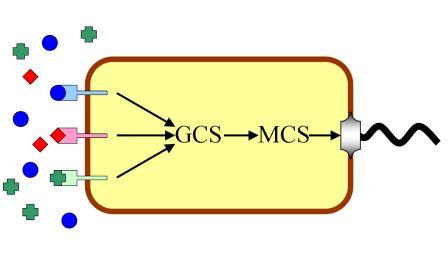
This year we are trying to work at the motility of bacteria. E.coli senses some chemicals and moves toward or stays still, this behavior is called chemotaxis. Any modification of the pathway from the sensor to the motile apparatus will cause different chemotactic effcts.
Moreover,most of the biosensors today are designed for a specific target signal, and they can not substitute each other in most cases. By structural analysis we find that most receptors for the same type of ligands exhibit great similarity, both in sequence information and characteristics. Our project is designed for an "Interchangeable Biosensor". The strategy is almost trial and error, to find an appropriate pathway between chemical molecules and corresponding reactions. Signals other than chemicals are much harder to manipulate, and integrating all the signals seems even more challenging,we are trying to isolate the elements and standardize them.
Part 1 Experimental Pathway
Part 2 The Experiments
Chemotaxis Project (1)1. Reconstruct pUC18 plasmid, insert GFP to the downstream of lac promoter
1.1 Use touchdown PCR to amplify GFP and add degradation tag and terminator. Primers: Forward: 5’- GA GAATTC G AGCAAGGGCGAGGAGCTGTTCACCG -3’
EcoR I
Reverse: 5’- CTTG CCCGGG TTATCACTTGTACAGCTCGTCCATGC -3’
Xma I
Template: provide by Guoqiang Chen’s lab in Tsinghua University 1.2 Cut pUC18 plasmid with EcoR I and KpnI. Purify the cut pUC18 from agarose gel with Kit. 1.3 Purify the PCR product GFP from agarose gel with Kit and cut with EcoR I and Kpn I. 1.4 Purify the cut GFP with column. 1.5 Run a gel to compare the concentrations of the fragment and vector in order to decide their volume in the ligation mixture. 1.6 Incubate the ligation mixture at 16 degree. 1.7 Use 10μl ligation mixture to transform 200μl DH5αcompetent cell, spread on the LB plate(Amp), and incubate at 37 degree over night. 1.8 Pick three clone to 5ml LB and shake at 37 degree for 12h, miniprep the reconstruct plasmid. 1.9 We sequenced the inserted part and it was right. 2. Synthesize the Nde I-Pm(complementary chain)-Pst I-Xba I-cad-Spe I-RBS(strong)-CI+LVA tag-Sac II-T1, T2-BamH I (the Blue part is the synthesis sequence) sequence. We use synthesis because we cannot use PCR to get the sequence from Staphyloccocus aureus Rosenbach 3. Insert CheZ(with tag and terminator) into the reconstruct pUC18(with GFP) 3.1 Extract the genome of E.coli DH5αstrain and use PCR amplify CheZ from it. Primer: Forward primer: 5’-GTCATGCC CA TATG CAACCATCAATCAAACCTGC-3’ Reverse primer: 5’-AT AGG CCT AAATCCAAGACTATCCAACAAATCGTCCACCTGATC-3’ 3.2 Purify the PCR product from gel and then cut with Nde I 3.3 Cut the pAcYc duet-1 plasmid with EcoR V and Nde I 3.4 Run a gel to compare the concentration of the vector and insertion in order to decide the their volumes in the ligation mixture 3.5 Ligate the vector and the cheZ, transformate DH5α and spread the LB plate(Chl). 3.6 Pick three clones to 5ml LB(Chl) and shake at 37 for 12h, then miniprep the reconstruct the plasmid. 3.7 Synthesize the Stu I-tag-Hind III-T0/T1T2-AatII sequence (with stick end), which contains CheZ’s tag and terminator. We firstly synthesized the sequence as two single strings, and then annealing them together as a double string DNA. The two single strings’ sequence is below: Forward 5’-CCT GCTGCAAACGACGAAAACTACGCTTTAGTAGCT TAA A AGCTT Stu I end LVA tag stop codon Hind III CGCAAAAAACCCCGCTTCGGCGGGGTTTTTTCGC GACGT-3’ T0 Aat II end Reverse 5’- C GCGAAAAAACCCCGCCGAAGCGGGGTTTTTTGCG A AGCTT TTA Aat II end T0 Hind III stop codon (complementary) AGCTACTAAAGCGTAGTTTTCGTCGTTTGCAGC AGG
LVA tag (complementary) Stu I end
3.8 Cut the pAcYcduet-1-cheZ plasmid with StuI and Aat II and column purify the vector 3.9 Run a gel to compare the concentration of the vector(pAcYcduet-1-cheZ) and insertion (Stu I-tag-Hind III-T0/T1T2-AatII) to make sure their volumes in the ligation mixture. 3.10 Ligate the two parts transformation spread plate pick 4 clones shake and get the reconstruct plasmid 3.11 We sequenced it and it was right. 3.12 Then we cut the whole CheZ-tag-terminator with NdeI and AatII and ligate the fragment into the pUC18-GFP vector, which is cut by the same two enzymes. 3.13 This sequenced this plasmid and the sequence was right. 4. Cut the synthesized Nde I-Pm(complementary chain)-Pst I-Xba I-cad-Spe I-RBS(strong)-CI+LVA tag-Sac II-T1, T2-BamH I (the Blue part is the synthesis sequence) sequence with BamH I and Nde I, the same was done on the vector(pUC18-cheZ-tag-ter-GFP). Then ligate them together and got a new reconstruct plasmid. It was also sequenced. 5. Put RBS-CI-tag into last vector. 5.1 Use PCR to amplify CI from lambda DNA, adding RBS to 5’ and tag to 3’ First round primers: 5’-GAGGGGACAAactagt ATGAGCACAAAAAAGAAACCATTAACACAAGAGC-3’
50 nt Tm=59.8
5’-TCGTTTGCTGCAGGCCT gccaaacgtctcttcaggccactgac-3’
43 nt Tm=63.5
Second round primers: 5’-C A CTAGT tctagaGAAAGAGGGGACAAactagt ATGAGCACAAAAAAGAAACC-3’
SpeI RBS 53nt Tm=49.6,65.0
5’-ACT CCGC GG TTA AGCTGCTAAAGCGTAGTTTTCGTCGTTTGCTGCAGGCCT gccaaacgtctcttcagg
Sac II LAA tag(complementary) 69nt Tm=53.4, >75
We also did a point mutation with PCR to mutate a Hind III restriction cite. For: 5’-GGCTCCAAGCCTAGCTTTCCTGACGGAATG-3’
30 nt Tm=65.6
Rev 5’- cattccgtcaggaaagctaggcttggagcc-3’ 5.2 Cut the RBS-CI-tag and the reconstructed vector with Sac II and Spe I and ligate them together. 5.3 Transformation the final plasmid to the ΔCheZ E.coli stain.
Now we have got the reconstructed bacteria!!!!!
Part 3
== Results ==
Simulation
Reference
1 Assessment of heavy metal bioavailability in contaminated sediments and soils using green fluorescent protein-based bacterial biosensors. Vivian Hsiu-Chuan Liao*, Ming-Te Chien, Yuen-Yi Tseng, Kun-Lin Ou. Environmental Pollution 142 (2006) 17e23
This literature helped us to introduce GFP into our project and gave us a description of heavy metal pollutants.
2 The C-terminal domain of the Escherichia coli RNA polymerase a subunit plays a role in the CI-dependent activation of the bacteriophage lambda pM promoter. Barbara Kedzierska , Anna Szambowska , Anna Herman-Antosiewicz , David J. Lee , Stephen J.W. Busby2, Grzegorz Wegrzyn1 and Mark S. Thomas3,* Nucleic Acids Research, 2007, Vol. 35, No. 7
This literature gives out a mechanism of CI-Pm promotor interaction.
3 Guiding Bacteria with Small Molecules and RNA.Shana Topp and Justin P. Gallivan. J. AM. CHEM. SOC. 2007, 129, 6807-6811 9
This literature lent us an overall vision of how bacteria movement happens and indicated out the central role of CheZ. It also introduced riboswitch into our project.
4 Luminescent bacterial sensor for cadmium and lead.Sisko Tauriainen, Matti Karp, Wei Chang, Marko Virta.Biosensors & Bioelectronics 13 (1998) 931–938
This literature descripted sensors for cadmium and lead.
5 Online and in situ monitoring of environmental pollutants: electrochemical biosensing of cadmium.I Biran, R Babai, K Levcov, J Rishpon, EZ Ron.Environmental Microbiology, 2000
This literature gives out a sensing and detecting method for cadmium.
6 Sensitivity of OR in Phage lambda.Audun Bakk, Ralf Metzler, and Kim Sneppen.Biophysical Journal Volume 86 January 2004 58–66
This literature explains the operation of operons in phage lambda.
| HOME | Team | Project | Parts | Modelling | Notebook | Doodle Board |
|---|
 "
"