Team:UC Berkeley Tools/Project
From 2008.igem.org
(→Clotho Development) |
(→Clotho Development) |
||
| Line 114: | Line 114: | ||
== Clotho Development == | == Clotho Development == | ||
| - | Ongoing | + | Ongoing development information on Clotho can be found [http://biocad-server.eecs.berkeley.edu/wiki/index.php/Clotho_Development here]. |
Revision as of 03:52, 7 August 2008
Contents |
Project Abstract
With >700 available parts and >2000 defined parts in the MIT Registry of Standard Biological Parts, and doubtless more parts local to the lab, synthetic biologists are now faced with the challenge of organizing, sorting, and editing these parts from an ever-growing collection of DNA. Due to this rapidly expanding collection, synthetic biology is reaching the point at which composite systems are becoming more and more prevalent. Computational tools are becoming increasingly needed to assist biologists to do the job, and it is no secret that many tools are indeed wheeling out into the community to help serve these needs.
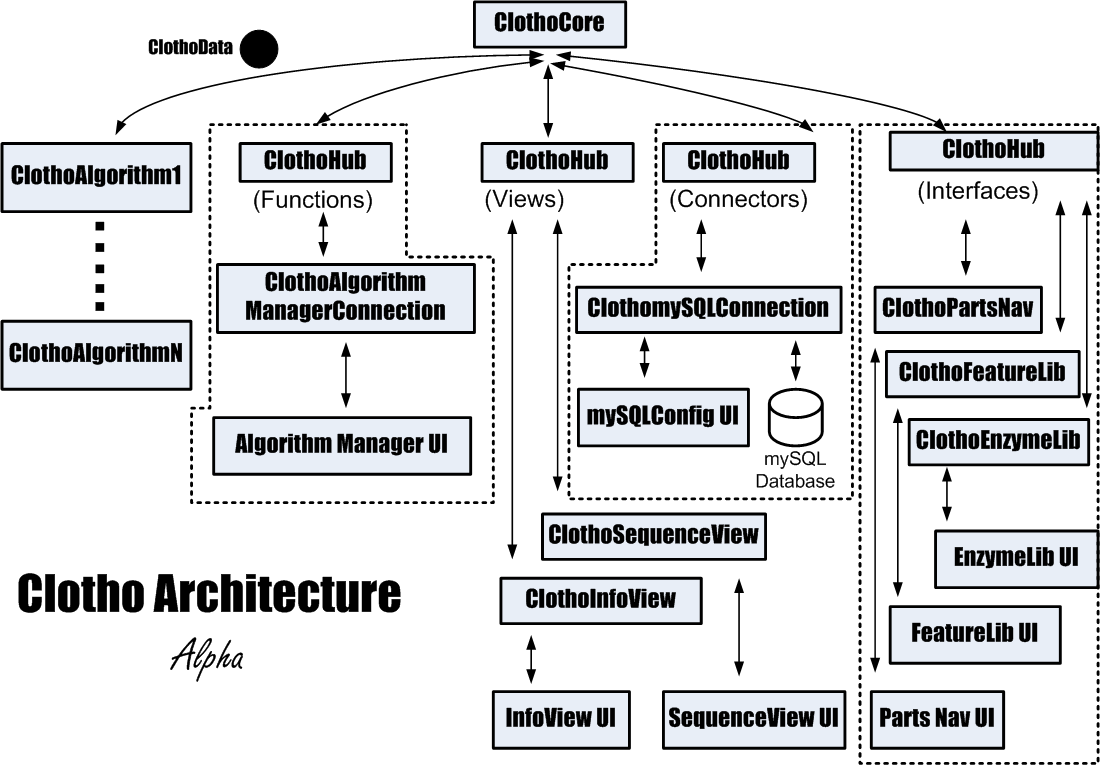
Clotho is a toolset designed in a structural paradigm to consolidate all these tools into one working, integrated toolbox. Clotho is based on a core-and-hub system which manages multiple connections, each connection serving an independent purpose in an almost self-sufficient manner. Examples of connections include sequence viewers/editors, parts database managers, sequence alignment GUIs, and much more. Computer-savvy biologists can create new connections which will then be integrated into Clotho. In this tool-consolidation and customization it is hoped that, in the future, biologists need not look further for a program that provides the answer to their needs.
Overall project
Clotho is based on a core-and-hub system which manages multiple connections, in which each connection serves an independent purpose in a self-sufficient manner. For instance, while one connection may be in charge of viewing/editing a sequence in an ApE-based manner, another connection may connect to databases, and will allow the user to receive and submit parts. Each connection may also perform more integrative tasks by passing data to each other through the core. If a user were working through the sequence view and database manager, for example, then the two connections could talk to each other if the user wished to edit a part in the sequence view and then resubmit the part back to the database.
Clotho's development page provides information on all current viewable connections.
Project Details
Overall Timeline
- iGEM 1st Meeting: June 2, 2008 - Nade and Matt start.
- Anne arrives: June 9th
- Check-up with Prof. Anderson: June 16, 2008
- iGEM picture session: July 7, 2008
- Clotho Testing Session 1: July 14, 2008
- Clotho Testing Session 2: July 16, 2008
- Clotho Alpha Release: July 26, 2008 - Download Here.
- Anne leaves (and sadness ensues): August 1, 2008
- Clotho Beta Release:
- iGEM 2008 Jamboree:
Getting Acquainted To Clotho
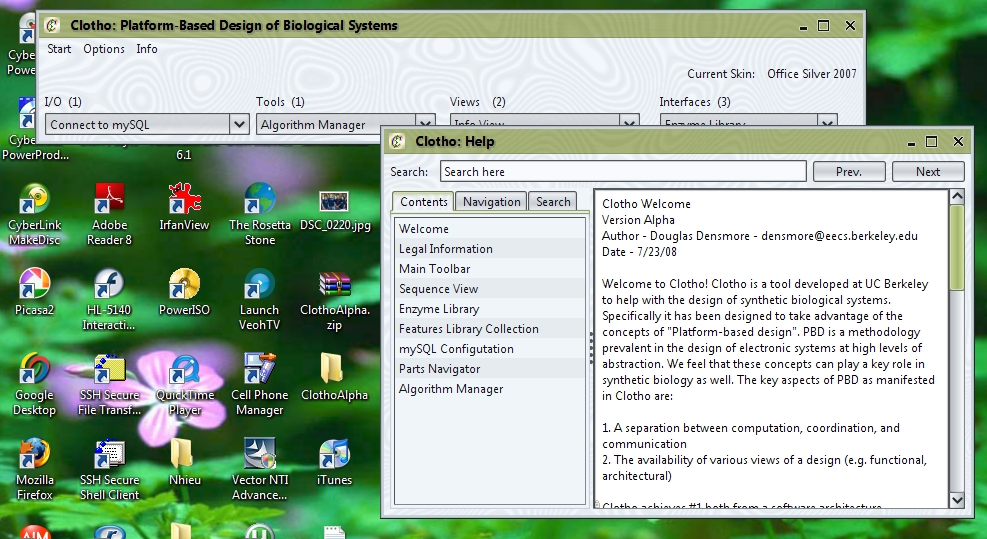
When the user first starts up the .jar file, they will notice that they are first greeted with Clotho's splash screen, and then the Clotho toolbar. The toolbar is the main window that will be used to access all connections from a central interface. Along the top of the toolbar, users can change the skins of the program by accessing Options->Set Skin. Under Info->Help, users can access documentation about various connections.
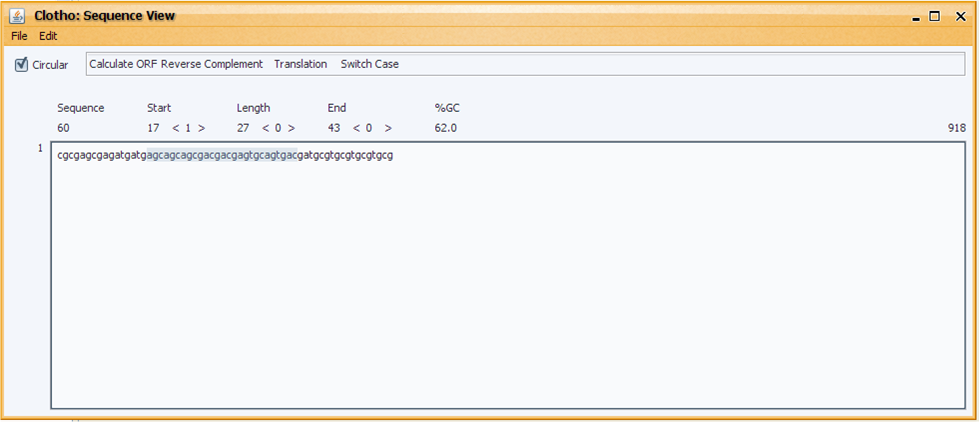
First-time users may be most comfortable with the sequence view before accessing any of the other connections. To access the sequence view, open Views->Sequence View.
Here users may view and edit sequences in the Fasta, Genbank, Strider and ApE formats. More information about the sequence view can be found under the sequence view help article.
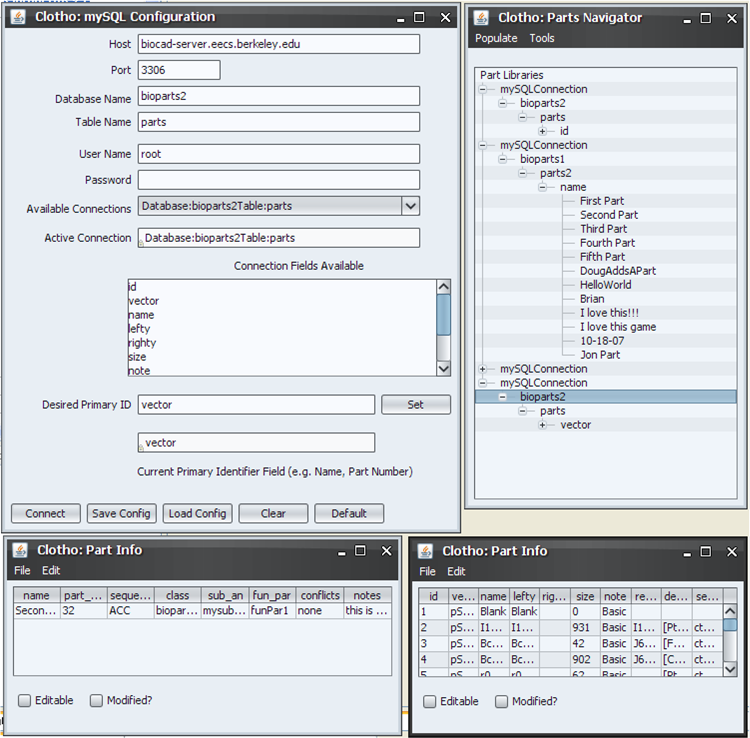
Users should also get acquainted to the Algorithm Manager and mySQL Connections. Return to the toolbar, open I/O->Connect to mySQL.
Here, users can interact with databases and sort through parts organized in mySQL. Once again, more information can be found in the corresponding help article. Also open here is the parts navigator, which is, for the time being, useful insofar as Clotho is connected to a mySQL database.
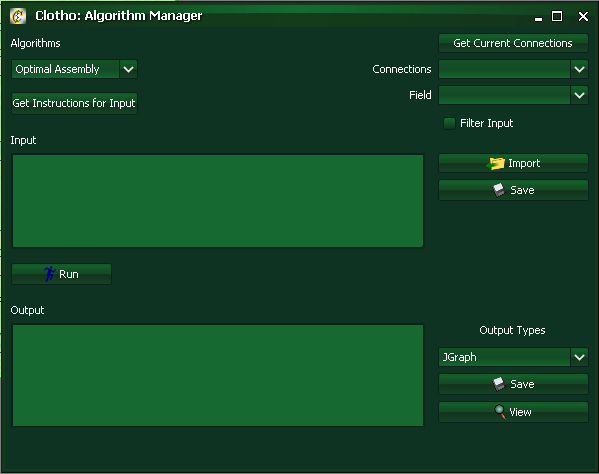
To access the Algorithm Manager Connection open Interfaces->Algorithm Manager.
The algorithm manager allows users tremendous creative control. The algorithm manager can take in an input, and spit out an output depending on the current algorithm implemented. Examples of possible algorithms include optimal assembly algorithms (look at 2ab assembly under the algorithm manager as a concrete example that also considers antibiotics), sequence alignment GUIs, and a icon viewer that can takes oligos and visually represents them in a graphical format. The possibilities here are near endless.
The Testing Sessions
Information and pictures regarding the testing sessions can be found here.
The Alpha Release
The Path Forward
Clotho Development
Ongoing development information on Clotho can be found here.
 "
"