|
|
| (25 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | | | |
| | {{Template:ClothoMenu}} | | {{Template:ClothoMenu}} |
| - |
| |
| | | | |
| | {{Template:ClothoTutorialVerticalMenu}} | | {{Template:ClothoTutorialVerticalMenu}} |
| | + | |
| | + | <!--YouTube--> |
| | + | <!---<html> |
| | + | <div align="right" class="youtube"> |
| | + | <object width="425" height="350"> <param name="movie" value="http://www.youtube.com/v/ktyt9mTOKIA"> </param> <embed src="http://www.youtube.com/v/ktyt9mTOKIA" type="application/x-shockwave-flash" width="425" height="350"> </embed> </object> |
| | + | </div> |
| | + | </html>---> |
| | + | |
| | + | |
| | | | |
| | <div id="mainContent"> | | <div id="mainContent"> |
| - | <p align="center" class="style1">Welcome to Clotho!<br />
| |
| - | </p>
| |
| | <p> </p> | | <p> </p> |
| | + | <p>'''<<< Previous''' | [[Team:UC_Berkeley_Tools/Project/Tutorial/Preferences|'''Next >>>''']]</p> |
| | <p><strong>1) The Main Toolbar</strong><br /> | | <p><strong>1) The Main Toolbar</strong><br /> |
| | This is the center of Clotho: the main toolbar. From here, you can open up any of the Clotho connections. The connections are separated into four categories: I/O, Tools, View, and Interfaces. "I/O" contains the connections that link Clotho to outside resources and databases. "Tools" contains the connections that provide functionality to Clotho. "Views" contains ways of viewing and manipulating information, including sequence data. "Interfaces" contains the connections that provide the user a way of interfacing with data, such as libraries of parts and feature annotations. From here, you can also access the Start, Help, and Options menus. For now, let's open up the options menu and select "Preferences"</p> | | This is the center of Clotho: the main toolbar. From here, you can open up any of the Clotho connections. The connections are separated into four categories: I/O, Tools, View, and Interfaces. "I/O" contains the connections that link Clotho to outside resources and databases. "Tools" contains the connections that provide functionality to Clotho. "Views" contains ways of viewing and manipulating information, including sequence data. "Interfaces" contains the connections that provide the user a way of interfacing with data, such as libraries of parts and feature annotations. From here, you can also access the Start, Help, and Options menus. For now, let's open up the options menu and select "Preferences"</p> |
| | [[Image:Tut1.PNG|center]] <p> </p> | | [[Image:Tut1.PNG|center]] <p> </p> |
| | + | </div> |
| | | | |
| - | <p><strong>2) Preferences</strong><br />
| + | '''<<< Previous''' | [[Team:UC_Berkeley_Tools/Project/Tutorial/Preferences|'''Next >>>''']] |
| - | In the preferences menu, you can set a number of options that affect various core Clotho components. Many of these options are used to specify the default locations for files used by other connections, but a few are used to change the behavior of these connections; we will come back to these later. Now, look under the "Look and Feel Settings" and select 'Office Blue 2007' from the Default Skin drop down menu - this sets it as teh default skin that Clotho will load up with. Now hit the "Apply Changes" button at the bottom of the Preferences window, and select 'Yes' when it asks if you want to overwrite the old preferences file. (You can also change the skin just for this session at any time in the Options menu of the main toolbar, under "Set Skin"). You should be looking at the main toolbar again, with a new look and feel.</p>
| + | |
| - | [[Image:Tut2.PNG|center]] <p> </p>
| + | |
| - | [[Image:Tut3a.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p><strong>3) Sequence View I</strong><br />
| + | |
| - | Now that we've got it everything looking spiffy, let's try out one of the main view of the tool - the Sequence View. On the main toolbar select "Sequence View" from the View drop down menu. This should bring up the Sequence View window. From here you have many options - you can load a sequence from a GenBank or FASTA file, drag in a sequence from another connection, copy and paste your chosen sequence, or enter it manually. Let's try opening a GenBank file ([http://openwetware.org/images/3/39/JCAseq_pSB1A2-Bca9128.str like this one], for example). Go to File, and select 'Load Sequence File' - this should bring up a window allowing you to choose what file you want to open. Go to the directory you saved the file to, select it, and click open (if the directory has many files in it, you can go to the 'Files of Type' drop down menu and filter it to only show GenBank or FASTA files). This will load the sequence into the current Sequence View window. From here, you can edit the sequence or perform a number of operations on it. But first, let's try adding some informative highlights to this plain text sequence. Go to the Highlighting menu, and select "Open Feature Library Connection".</p>
| + | |
| - | [[Image:Tut3b.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p><strong>4) Feature Highlighting</strong><br />
| + | |
| - | The Features Library manages the local libraries of feature annotations for sequences. From here you can create, load, or remove libraries, examine features from within these libraries, and select which libraries you want to use for highlighting. (Additionally, you can drag features from a library into the Sequence View and place them anywhere in the sequence.) For now, let's select default library and hit the "Highlight" button. Close the Feature Library and go back to the Sequence View, and you'll see that several features have been highlighted in different colors. If you run the mouse cursor over any of the highlights, the name of the feature will pop-up on the info bar just above the sequence box. Knowing where our features of interest are helps, but let's find some restriction sites where we might want to cut our sequence. Go to the Highlighting menu, and select "Open Enzyme Library".</p>
| + | |
| - | [[Image:Tut4a.PNG|center]] <p> </p> | + | |
| - | [[Image:Tut4b.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p><strong>5) Restriction Site Highlighting</strong><br />
| + | |
| - | The Enzyme Library contains a list of restriction enzymes that you can search for in the sequence. You can select each enzyme individually, search for them by name or sequence, or select a pre-assigned group of enzymes. Let's try selecting the standard four BioBrick enzymes [EcoRI, PstI, SpeI, and XbaI]: you can do this by selecting them manually or by going to "Select by Group" and picking the "BBa Enzymes" group. Select the "Highlight Enzymes" button, and open the Sequence View window, but don't close the Enzyme Library just yet. You should see new orange highlights in the sequence corresponding to the selected restriction, and just like features, running the mouse cursor over the highlights shows the corresponding restriction enzyme. [Clotho supports multiple highlighting, so you may see features and restriction sequences overlapping one another, and in that case both names will be shown].</p>
| + | |
| - | [[Image:Tut5a.PNG|center]] <p> </p>
| + | |
| - | [[Image:Tut5b.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p><strong>6) Sequence View II</strong><br />
| + | |
| - | Now let's play with some of the other basic tools in the Sequence View. Try pressing First, let's find an open reading frame - place your cursor anywhere in the sequence, go to the "ORFs" menu and select "Find Next ORF". Alternatively, you can use the shortcut and press [Ctrl + .]. This will place line a light green line underneath the next open reading frame found in the sequence after the cursor (if other alternate start codons are enabled in the preferences, their reading frames will be shown using yellow or red lines). Highlight this sequence with the mouse and select the "Translate" option from the "Tools" menu (or press [Ctrl + T]). The protein translation of this reading frame will be printed to the 'Output Data' window. Similiarly, if you select "Reverse Complement" [Ctrl + /], the reverse complement of the sequence will be printed. "Find Reverse ORF" and "Reverse Translate" operate in the same fashion, but they look at the reverse complement of the sequence. "UPPER CASE", "Switch Case", and "lower case" all operate on any portion of the sequence you care to highlight (case has no effect on sequence analysis and is treated as aesthetic). Highlighting a string of nucleotides also displays information like G-C content and melting temperature. Additionally, you can open up a tools menu under "Tools" [Ctrl + F] to peform find and replace operations on the sequence. Let's leave the Sequnce View for now - either select "Exit Sequence View" from the File menu or click the 'X' in the corner and select "Yes close" from the resulting pop-up box.</p>
| + | |
| - | [[Image:Tut6.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p><strong>7) Connecting to a Database</strong></p>
| + | |
| - | [[Image:Tut7.PNG|right]]
| + | |
| - | <p>Some of Clotho's most interesting features can only be utilized once we've connected to a database. Let's try setting up a mySQL connection: on the Main Toolbar select "PoBoL based mySQL" from the I/O drop down menu. This should bring up the mySQL/PoBoL Configuration window. The first two boxes are username and password, but don't put anything in them just yet. First, select the "Create New Connection" tab, and enter the following information:</p>
| + | |
| - | <p>Name:<br />
| + | |
| - | Host:<br />
| + | |
| - | Port:<br />
| + | |
| - | Database:</p>
| + | |
| - | <p>After you're done, click "Create" and go to the "Existing Connections" tab. Select the connection you just made from the "Connection Available" drop down menu. In the appropriate boxes, put in the username **USERNAME** and password **PASSWORD**. Then click "Set as Next, we'll setup the PoBoL binding file: click the "View PoBoL Binding" button, and it should bring up the PoBoL Binding window.</p>
| + | |
| - | <p> </p>
| + | |
| - | | + | |
| - | <p><strong>8) PoBoL</strong></p>
| + | |
| - | <p>PoBoL stands for "Provisional Biobrick Language", and represents a way to translate different databases into one common language. The binding file is a way of specifying which tables and entries from the database correspond to which PoBoL data types. Ideally, you would be able to download a binding file for any given database, saving you the trouble - for our tutorial database, such a file can be found **HERE** - just copy and paste the contents of the text file into the 'Binding Configuration' window. If no such file existed though, you would have to create it manually: first you would select a Table and then a Field from the 'Database Information' box, and then a corresponding Object and Field from the 'PoBoL fields' box, and click "Bind". This would have to be done for all the tables and fields you wish to use from the database. Fortunately, you only have to create the mySQL connection and its corresponding PoBoL binding file once. Now click "Save/Connect", and select 'Yes' when the message box appears. Now Clotho is connected! (Note: In future uses, you will only have to enter the user name and password and click "Connect".)</p>
| + | |
| - | <p>More information on PoBoL can be found **HERE**.</p>
| + | |
| - | [[Image:Tut8.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p><strong>9) Parts Manager and Plate Manager</strong></p>
| + | |
| - | <p>Two Clotho connections so far utilize the PoBoL-based mySQL connection: the Parts Manager and the Plate Manager. On the Main Toolbar select "Parts Manager" from the Interfaces drop down menu, bring up the Parts Manager window. The Parts Manager allows you to look at BioBrick parts in the database. It contains a lot of information on each part, including the part and vector nicknames, the parts status, who submitted it, its location in the lab, the amount left, and more. In the future, you will be able to add, delete, and edit parts in the database through this window, as well as looking up more detailed information on the part.</p>
| + | |
| - | [[Image:Tut9a.PNG|center]]
| + | |
| - | <p>Close the Parts Manager, and on the Main Toolbar select "Plate Manager" from the Tools drop down menu, bringing up the Plate Manager window. The Plate Manager lets you keep track of the contents and locations of plates in the lab. From the drop down menu below the 'Zoom' buttons, select **PLATE NAME*, click the "Select" button, and select 'Yes' in the following pop-up box - this should load up a 96-well plate from the database. Information on the plate itself can be found in the 'Plate Information' box. Clicking on any of the occupied wells will fill out the 'Sample Information' box with information on the contents of the well. The data that gets displayed can be changed in the Plate Manager preferences window, by going to the Edit menu item, selecting "Preferences", and changing the the labels. For now, let's close the Plate Manager window and go back to the Main Toolbar.</p>
| + | |
| - | [[Image:Tut9b.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p><strong>10) Algorithm Manager</strong></p>
| + | |
| - | <p>From the Main Toolbar select "Algorithm Manager" from the Tools drop down menu. The Algorithm Manager is a straightforward but robust tool - you take some input, select an algorithm, and it gives you an output. Each algorithm has its own rules for its use, including the proper inputs and syntax. After selecting any algorithm from the 'Algorithms' drop down box, select "Get Instructions for Algorithm" to get information on how to get it up and running. After finding an appropriate input (whether entered manually, imported from a file, or exported from another connection) select the "Run" button, and the output should appear in the 'Output' box. From here you can view or save the information in a number of different formats, graphically or textually. The default Optimal Assembly and 2abAssembly algorithms provide methods for optimizing the number of steps required to create composite parts.</p>
| + | |
| - | [[Image:Tut10.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p><strong>11) Other Information</strong></p>
| + | |
| - | <p>From here, feel free to explore Clotho! More detailed information on the different connections can be found in the Help menu.</p>
| + | |
| - | <p> </p> [[Image:Tut11.PNG|center]] <p> </p>
| + | |
| - | | + | |
| - | <p> </p>
| + | |
| - | </div>
| + | |
<<< Previous | Next >>>
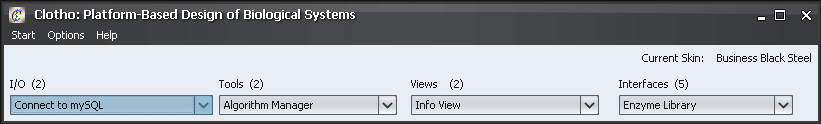
1) The Main Toolbar
This is the center of Clotho: the main toolbar. From here, you can open up any of the Clotho connections. The connections are separated into four categories: I/O, Tools, View, and Interfaces. "I/O" contains the connections that link Clotho to outside resources and databases. "Tools" contains the connections that provide functionality to Clotho. "Views" contains ways of viewing and manipulating information, including sequence data. "Interfaces" contains the connections that provide the user a way of interfacing with data, such as libraries of parts and feature annotations. From here, you can also access the Start, Help, and Options menus. For now, let's open up the options menu and select "Preferences"
 "
"