Team:University of Lethbridge/Notebook/Project2August
From 2008.igem.org
m |
m |
||
| Line 150: | Line 150: | ||
====Nathan Puhl, Roxanne, Andrew==== | ====Nathan Puhl, Roxanne, Andrew==== | ||
-Performed a colony PCR on representative colonies containing the pGEM T-easy plasmid to screen for the presence of the riboswitch. | -Performed a colony PCR on representative colonies containing the pGEM T-easy plasmid to screen for the presence of the riboswitch. | ||
| + | |||
| + | Same as previous protocol, However, Annealing Temperature is set to 65.0 C | ||
-Ran a gel of the PCR products on 3% Agarose @ 100V for 27 minutes. | -Ran a gel of the PCR products on 3% Agarose @ 100V for 27 minutes. | ||
| + | |||
| + | ===August 31, 2008=== | ||
| + | ====Roxanne==== | ||
| + | -Reran the gel of the colony PCR, determined that it did not work. | ||
| + | |||
| + | -Performed a second colony PCR on 3 white colonies, 1 blue colony and 1 blue/white colony from the pGEM T-easy + RS1/RS2 recombinant cells. | ||
| + | |||
| + | Same as Previous Protocol, annealing temperature is 50.0C | ||
| + | |||
| + | -Ran a 2% Agarose Gel of the PCR Products at 100V for 27 minutes. | ||
| + | |||
| + | -One of the White Colonies and the Blue/White Colony in each case amplified. Those colonies wer picked and incubated at 37.0C overnight. | ||
Revision as of 23:30, 3 September 2008
Back to The University of Lethbridge Main Notebook
Contents |
August 1, 2008
Nathan Puhl, Roxanne
Riboswitch 20 uL PCR. Set up 4 reactions (25 uL for each total volume). 1 uL or 1/100 pTopp and 1 uL or H1/100 PCR from July 28, 2008.
PCR conditions:
A. Initial denaturation: 98 C (3 min)
B. -Denaturation: 98 C (10 sec)
- Annealing: 55 C (30 sec)
-Extension: 72 C (15 sec)
-30 cycles
C. Final extension: 72 C (7 min)
August 2, 2008
Nathan Puhl, Roxanne
Ran riboswitch (Aug. 1, 2008) on 3% agarose gel. Results are in the hard copy lab notebook. Looks like 76 bp band will extract.
August 7, 2008
Nathan Puhl, Roxanne
Riboswitch:
Set up PCR using purified riboswitch from Aug. 5, 2008 with platinum Taq (50 uL reaction). Made Master Mix for three reactions. Master Mix:
-10x Buffer (no Mg2+): 15 uL -10 mM dNTPs: 3 uL -50 mM Mg2+: 4.5 uL -10uM RF: 3 uL -10uM RR: 3 uL -Plat. poly: 0.6 iL -H20: 120.9 uL -template: 1 uL
Cycle conditions:
A. Initial denaturation: 94 C (2 min)
B. -Denaturation: 94 C (30 sec)
- Annealing: 55 C (30 sec)
-Extension: 72 C (30 sec)
-30 cycles
C. Final extension: 72 C (7 min)
Roxanne
-Ran the PCR Product on a 3% gel using only 1uL of DNA from the riboswitch.
August 16, 2008
Nathan Puhl, Roxanne, Munima
-Restriction Digested the purified riboswitch and pSB1A7 with XbaI and SpeI, let it run for 4 hours
Nathan Puhl, Roxanne
-Ran all of the restricted pSB1A7 plasmid through a 1% agarose gel at 100V for 25 minutes. -Ran a gel extraction on the pSB1A7 cut plasmid, and ran a PCR clean-up reaction on the digested RS1 and RS2 amplicons. -Ran 1 uL of each on a 1% to quantify the amount of DNA present. -Ligated RS1 + pSB1A7, and RS2 + pSB1A7, using T4 DNA Ligase.
-1 uL of RS1/RS2 -4 uL of pSB1A7 -1 uL of 10x T4 DNA Ligase Buffer -0.33 uL of T4 DNA Ligase -3.67 uL of water
allowed the reaction to go overnight
August 17, 2008
Nathan Puhl
-Transformed DH5a cells with the pSB1A7 + RS1, and pSB1A7 + RS2 plasmids.
-Plated on semi-solid agar plates containing 100 ug/mL of ampicillin.
August 18
Christa, Nathan Puhl, Munima
-Nathan checked the plates for growth. Colonies are present.
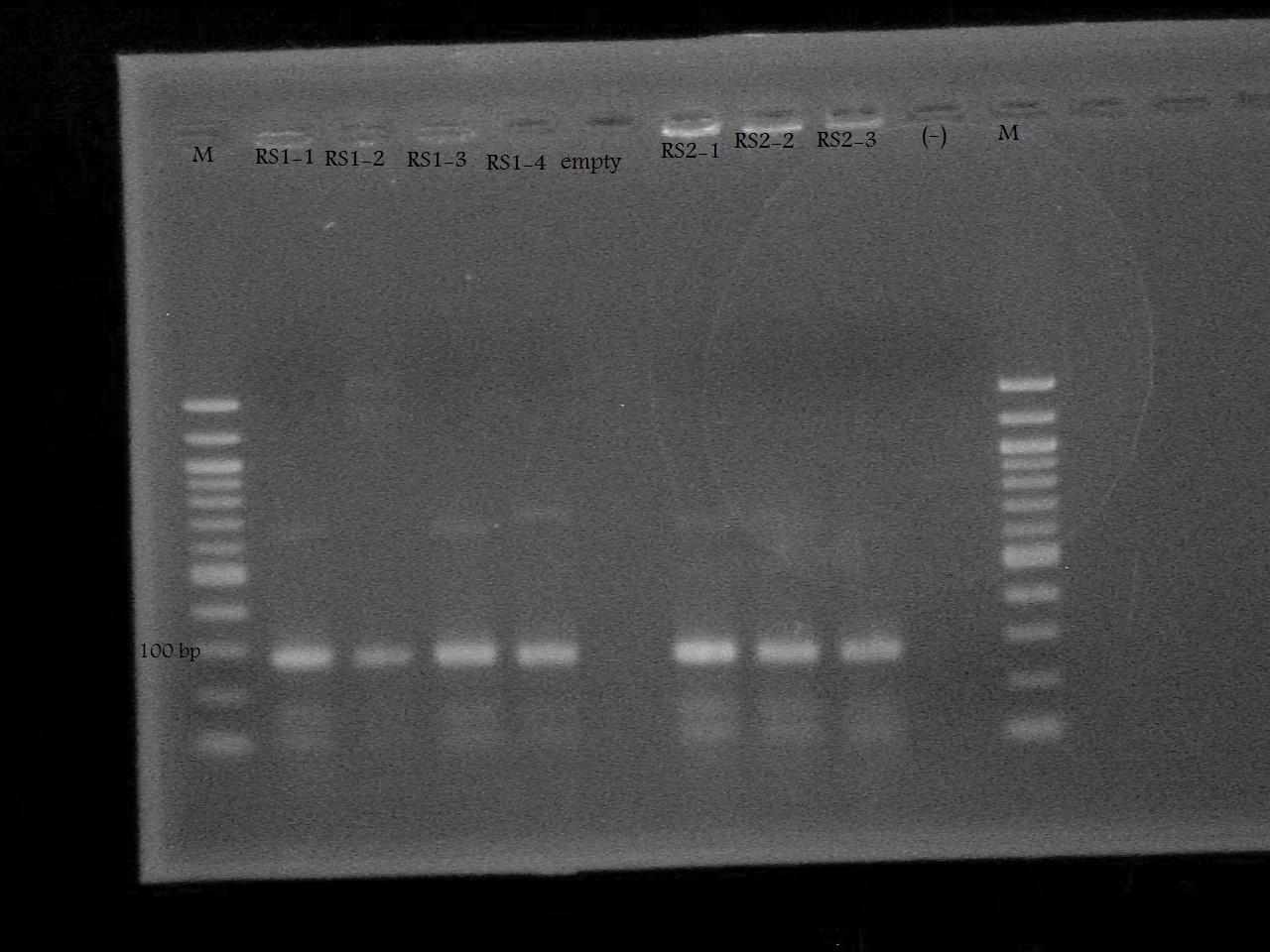
-Ran a colony PCR of the pSB1A7 + RS1, and pSB1A7 + RS2 recombinant cells transformed by Nathan and Roxanne on August 17.
-Inoculated the cells into tubes of liquid media + 100 ug/ml of ampicillin.
Roxanne will remove the PCR tube from the thermocycler in the morning.
August 19, 2008
Roxanne
-Ran the PCR Product on a 2% Agarose Gel at 100 V for 33 minutes.
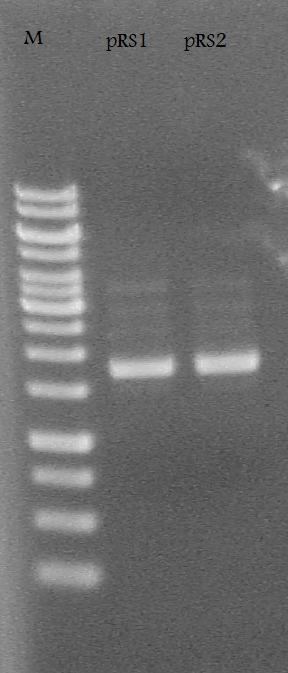
-Plasmid Prepped and made glycerol stocks from the RS1-1 and RS2-1 tubes of cells incubated in LB media + 100 ug/mL ampicillin.
Ran the pRS1 and pRS2 plasmids on a 1% Agarose Gel at 100 V for 30 minutes.
August 21
Nathan Puhl, Roxanne
-Screened the pSB1A7 + RS1, and pSB1A7 + RS2 by PCR using the VF2 and RS1/RS2 Reverse Primers determine whether the plasmids obtained from the recombinant cells contain the riboswitch, and if so, if it inserted in the correct orientation.
August 21, 2008
Roxanne
-Ran the PCR products on a 1% Agarose Gel at 100 V for 33 minutes. The gel was empty with the exception of primer dimers.
Nathan Puhl, Roxanne
-went over the SELEX protocol with HJ to determine the primers we will need to do this, and how exactly we plan on perfoming the evolution.
-setup a restriction digest for pSB1A7 using XbaI and SpeI, ran overnight.
August 23, 2008
Nathan Puhl, Roxanne
-Digested pSB1A7 with Antarctic Phosphatase
-9 uL of cut pSB1A7 -1.5 uL of 10x Antarctic Phosphatase Buffer -1 uL of Antarctic Phosphatase Enzyme -3.5 uL of water Allowed the Reaction to take place for 30 minutes to remove the 5` Phosphates from the pSB1A7 plasmid to prevent religation.
-Ran the remainder of the pSB1A7 plasmid from August 22nd on 1 1% Agarose Gel at 100 v for 27 minutes.
-Gel Extracted the plasmid DNA.
-Purified the Phosphatase reaction to isolate the pSB1A7 DNA.
-Ran a 1% gel to quantify the amount of plasmid DNA present.
-Ligated RS1 and RS2 into the dephosphorylated pSB1A7 using T4 DNA Ligase.
-1 uL of RS1 or RS2 -4 uL of dephosphorylated pSB1A7 -1 uL of 10X T4 DNA Ligase Buffer -0.33 uL T4 DNA Ligase Enzyme -3.67 uL water Reaction was allowed to go overnight
August 24, 2008
Nathan Puhl
-Transformed DH5a cells with the RS1+pSB1A7 or RS2+pSB1A7 plasmid on semi-solid agar plates containing 100 ug/mLof ampicillin.
August 29, 2008
Roxanne, Nathan Puhl, Munima, Sebastian, Andrew
-Performed the ligation of the riboswitch into pGEM T-easy, transformed and plated.
August 30, 2008
Nathan Puhl, Roxanne, Andrew
-Performed a colony PCR on representative colonies containing the pGEM T-easy plasmid to screen for the presence of the riboswitch.
Same as previous protocol, However, Annealing Temperature is set to 65.0 C
-Ran a gel of the PCR products on 3% Agarose @ 100V for 27 minutes.
August 31, 2008
Roxanne
-Reran the gel of the colony PCR, determined that it did not work.
-Performed a second colony PCR on 3 white colonies, 1 blue colony and 1 blue/white colony from the pGEM T-easy + RS1/RS2 recombinant cells.
Same as Previous Protocol, annealing temperature is 50.0C
-Ran a 2% Agarose Gel of the PCR Products at 100V for 27 minutes.
-One of the White Colonies and the Blue/White Colony in each case amplified. Those colonies wer picked and incubated at 37.0C overnight.
 "
"