Team:University of Lethbridge/Notebook/Project3October
From 2008.igem.org
Andrew.Field (Talk | contribs) m (→October 7 2008) |
Andrew.Field (Talk | contribs) m |
||
| Line 25: | Line 25: | ||
'''Andrew''' | '''Andrew''' | ||
| - | Ligation of +18 rpsA DNA into pGEM | + | Ligation of +18 rpsA DNA into pGEM T-easy beacuse primer design requires DNA to be in a plasmid for endonuclease activity of restriction enzymes. |
Heat deactivated at 65 C after 1 hour incubation at 37 C. | Heat deactivated at 65 C after 1 hour incubation at 37 C. | ||
| Line 35: | Line 35: | ||
'''Andrew''' | '''Andrew''' | ||
| - | Transformation of +18 rpsA TIR DNA into pGEM | + | Transformation of +18 rpsA TIR DNA into pGEM T-easy to make enough DNA to be restricted/ligated into biobrick format. (psB1A2) |
50 uL og xgal added to LB + amp plates to screen for blue and white colonies. | 50 uL og xgal added to LB + amp plates to screen for blue and white colonies. | ||
| Line 46: | Line 46: | ||
'''Andrew''' | '''Andrew''' | ||
| - | Transformation into pGEM | + | Transformation into pGEM T-easy successful, many white and some blue colonie on both plates. |
Picked 3 colonies from each plate to grow in 5mL LB + amp for a pDNA prep. | Picked 3 colonies from each plate to grow in 5mL LB + amp for a pDNA prep. | ||
Revision as of 01:44, 28 October 2008
Back to The University of Lethbridge Main Notebook
Contents |
October 7 2008
Andrew, Nathan Puhl
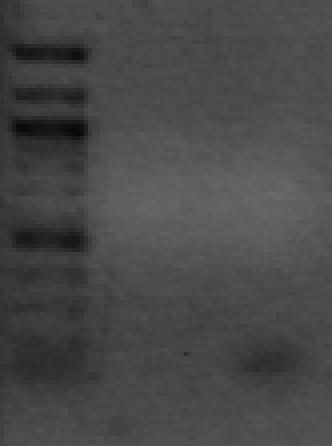
Ran a gel to confirm presence of DNA from PCR on September 30 2008. DNA present in all wells, so a 2% agaraose gel was prepared for a gel extraction of DNA. 40uL of DNA was loaded and gel extracted using a kit.
October 14 2008
Andrew
Ran 1uL of gel extraction products on a gel to determine presence/quantity of DNA.
Band close to ~150 bp for the +18 rpsA DNA, which is approximately the correct size. No band present for the +1 TIR DNA, meaning gel extraction was not successful.
October 16 2008
Andrew
Ligation of +18 rpsA DNA into pGEM T-easy beacuse primer design requires DNA to be in a plasmid for endonuclease activity of restriction enzymes.
Heat deactivated at 65 C after 1 hour incubation at 37 C.
October 21 2008
Andrew
Transformation of +18 rpsA TIR DNA into pGEM T-easy to make enough DNA to be restricted/ligated into biobrick format. (psB1A2)
50 uL og xgal added to LB + amp plates to screen for blue and white colonies.
October 22 2008
Andrew
Transformation into pGEM T-easy successful, many white and some blue colonie on both plates.
Picked 3 colonies from each plate to grow in 5mL LB + amp for a pDNA prep.
October 23 2008
Andrew
pDNA prep of 6 culture tubes using a kit
 "
"