Team:University of Ottawa/Project
From 2008.igem.org
(→Journal Club) |
(→Journal Club) |
||
| Line 100: | Line 100: | ||
=='''Journal Club'''== | =='''Journal Club'''== | ||
===PTP2 Overexpression=== | ===PTP2 Overexpression=== | ||
| - | + | Maeda T, Wurgler-Murphy S M, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242-245 | |
===Cytokinin=== | ===Cytokinin=== | ||
Werner T, Motyka V, Strnad M, Schmulling T (2001)Regulation of plant growth by cytokinin. PNAS 98 (18) 10487-10492 | Werner T, Motyka V, Strnad M, Schmulling T (2001)Regulation of plant growth by cytokinin. PNAS 98 (18) 10487-10492 | ||
Revision as of 19:26, 23 July 2008
Contents |
Project Overview
When large quantities of protein are required for industrial, pharmaceutical and research purposes, researchers may resort to expression of the loci of interest in a genetically modified organism. Once these transgenic organisms are built, they will be grown to sufficient titer before undergoing extraction for the protein of interest. In the case of microorganisms, this is performed in large bioreactors, where the protein of interest will be extracted from a continuously growing cell culture. When the protein of interest is excessively large, synthesis by the organism is difficult, and the use of bioreactors is limited. In the bioreactor, a small subpopulation of cells emerges, where the cells are not expressing the protein correctly. This subpopulation soon dominates due to positive selective pressure, and the use of the bioreactor becomes less effective with time. After a couple of months, the bioreactor will become inefficient, and will have to be “restarted”, a long and costly process. The goal of this year’s uOttawa iGEM team consists of generating a yeast strain capable of long term protein expression.
Pulsate Gene Expression
Gene expression in cells is a dynamic process in which transient responses are often observed. This can be conceptualized as a “pulse” of protein concentration. Achieving such a pulse involves a complex network of activities, which makes it difficult to elucidate the mechanisms in naturally occurring systems. Nevertheless, simple synthetic networks can be created to model this behaviour. We are working on a feed-forward regulatory motif to generate a pulsate expression of a protein of interest, in the model organism Saccharomyces cerevisiae. The general feed-forward mechanism is illustrated in Figure 1A. Upon stimulation (A), a combinatorial promoter is turned on, resulting in expression of the protein of interest (C). The stimulation also turns on another promoter encoding a repressor protein (B), which subsequently acts on the first combinatorial promoter to turn off expression of the protein of interest. Thus, a pulse in protein of interest concentration is achieved.
Cell-to-Cell Communication
An important aspect of naturally occurring biological systems is the cell-to-cell communication that triggers the biomolecular responses in multicellular settings. Our pulse generator system uses signalling elements from Arabidopsis thaliana that have been implemented in yeast [Ref: Chen & Weiss, 2005], to induce the feed-forward network. One population of cells send the signal molecule cytokinin isopentenyladenine (IP), while another population receives the signal via the cytokinin receptor AtCRE1. In combination, the engineered cells receive the IP signal, which induces the pulsate expression of the protein of interest.
Oscillatory Dynamics
After the pulsate protein expression event, the final state of the system consists of a repressed protein of interest. It is desirable to regenerate the system so that it is sensitive to the provided stimulus again – i.e., so that the protein of interest is not repressed. This can be done by destruction of the original inducing signal molecule. Cytokinin dehydrogenase is an enzyme from A. thaliana that degrades the IP signal molecule, and has also been implemented in S. cerevisiae [Ref: Frebortova et al., 2007]. This enzyme is added to the feed-forward network in the manner illustrated in Figure II. Cytokinin dehydrogenase (D) is turned on slowly, and eventually inhibits the stimulus signal (A). With the signal degraded, the repressor protein (B) is no longer produced. With normal protein, any repressor protein present in the cell will inevitably be degraded. Finally, with neither the signal (A) nor the repressor (B) present, the system returns to its initial state.The two populations of sender and receiver cells can be grown together. The inducing IP signal is continually supplied to the receiver cells, which would in turn go through cycles of IP stimulation and degradation. Thus oscillatory dynamics of pulsate protein expression can be accomplished.
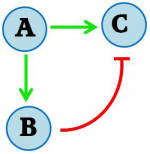
Figure I - General model of the feed forward regulatory motif
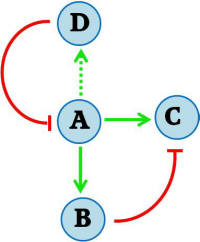
Figure II - An additional element is added to the feed forward regulatory motif, where protein D inhibits the initial stimulus A
Applications
With our pulse generator protein expression system, the proteins are only produced in short pulses, reducing the burden on the cells, and preventing toxic accumulation. These engineered cells can thus be resilient in a bioreactor setting, making them ideal for large-scale protein production applications.
Journal Club
PTP2 Overexpression
Maeda T, Wurgler-Murphy S M, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242-245
Cytokinin
Werner T, Motyka V, Strnad M, Schmulling T (2001)Regulation of plant growth by cytokinin. PNAS 98 (18) 10487-10492
Cytokinin Dehydrogenase
Schmulling T, Werner T, Riefler M, Krupkova E, Bartrina y Manns I (2003) Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res 116:241-252
Frebortova J, Galuszka P, Werner T, Schmulling T, FrebortI (2007) Functional expression and purification of cytokinin dehydrogenase from Arabidopsis thaliana (AtCKX2) in Saccharomyces cerevisiae. Biologia Plantarium 51 (4):673-682
 "
"