Team:University of Ottawa/Project
From 2008.igem.org
The Pulsilator
A synthetic pulse-generator in yeast for sustainable expression of recombinant proteins
Large-scale production of recombinant proteins typically involves growing genetically modified microorganisms in bioreactors in which metabolic stress and selective pressure tends to diminish culture productivity over time. To alleviate the detrimental effects of continual high-rate synthesis, we have designed a yeast strain capable of producing bursts of gene expression in a controlled and inducible manner. These "pulses" are generated by the action of an inducer molecule that triggers the synthesis of a protein of interest and simultaneously induces a repressor protein to terminate expression as well as an enzyme to degrade the inducer signal, thereby returning the system to its initial state. By co-culturing populations of inducer-synthesizing cells and pulse-generating receiver cells, we hope to achieve self-sustaining oscillatory gene expression dynamics that could render long term culture-based recombinant protein synthesis more sustainable. This would open the door for the production of considerably toxic proteins for numerous applications including anti-cancer therapeutics and antiseptics.
The problem
...
Yeast
...
The template
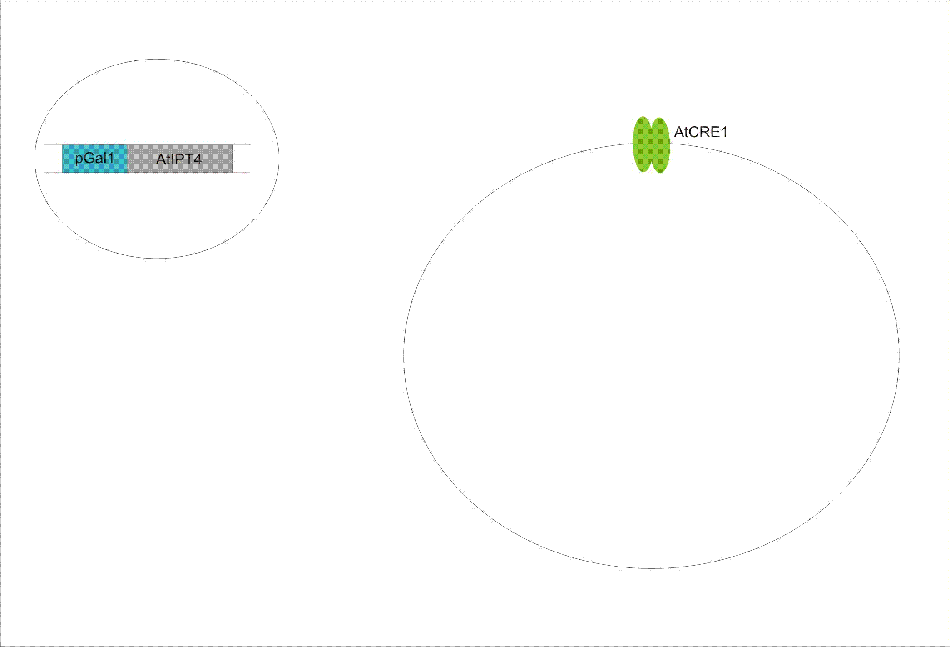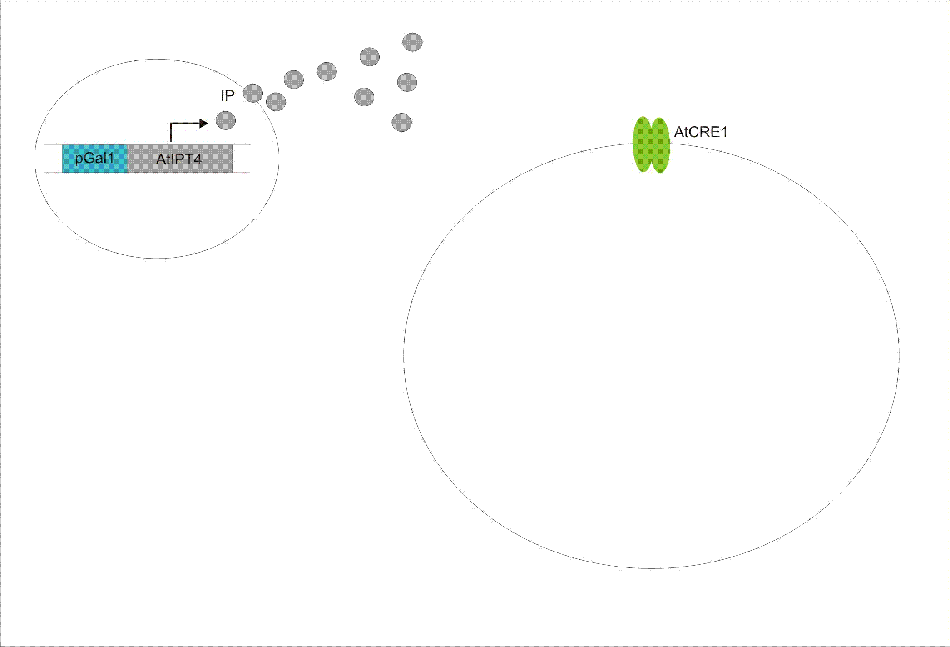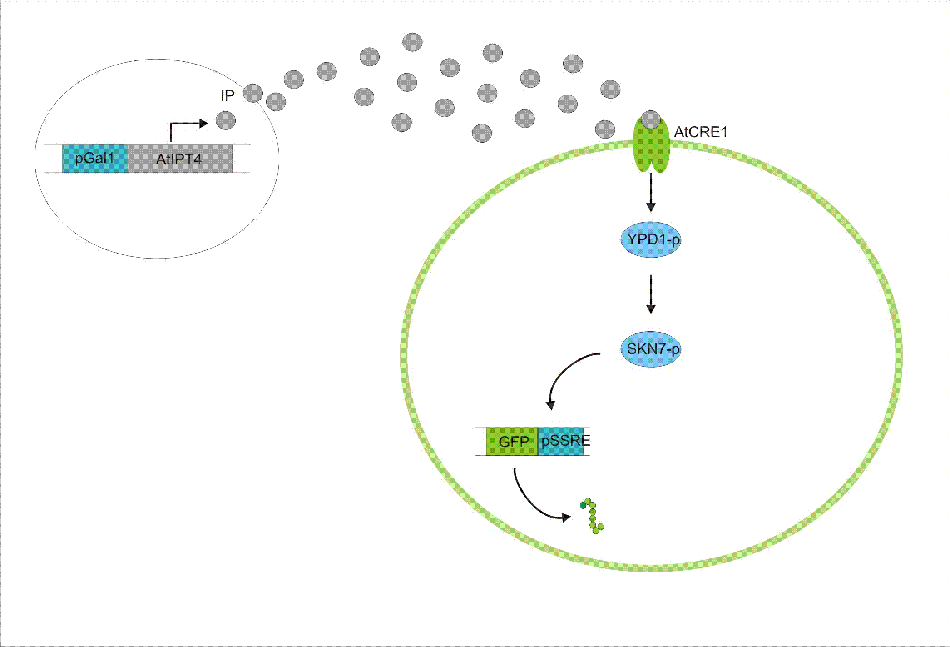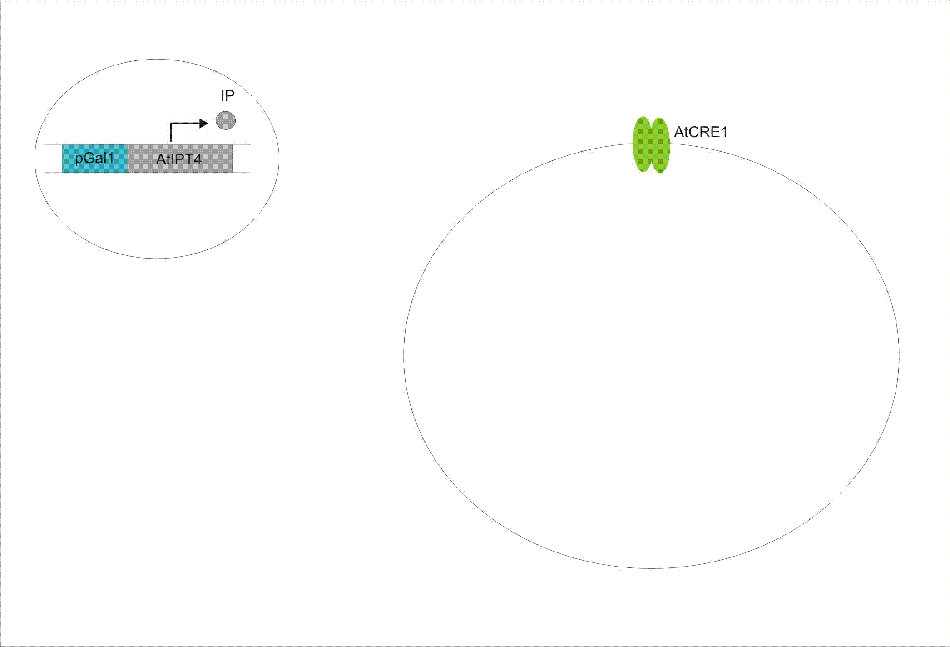
We discovered a useful template system developed by Ming-Tang Chen and Ron Weiss of Princeton University. They developed a yeast strain containing signaling elements from the flowering plant, Arabidopsis thaliana. These yeast express a surface cell receptor AtCRE1 that binds a cytokinin, a type of plant hormone, called isopentenyladenine (IP). This signaling element triggers an endogenous signal transduction pathway , leading to the activation of the nuclear aspartate response regulator SKN7, a transcription factor. By employing a synthetic promoter constructed from a modified yeast MEL1 promoter and containing one or two tandem synthetic SKN7 response elements (SSRE), target genes can be put under inducible control of IP. There are several advantages to using such an inducer molecule. Being completely foreign to the host organism reduces the concern of affecting other pathways or of endogenous activation of the pathway. IP is relatively cheap compared to other inducer compounds and can be synthesized on site using the recombinant enzyme from A. Thaliana, AtIPT4. Our ultimate plan is to incorporate this enzyme into a strain of sender cells as part of a dual population system. In addition, given that the selection of biobricks for yeast is currently limited when compared to the spectrum of parts developed for E. coli, we thought that this would be a novel yeast-based system to expand upon.
The original CRE1 system
To construct the original yeast strain, several genetic manipulations had to be performed to ensure proper function. It is known that upon binding inducer CRE1 phosphorylates the histidine phosphor-transfer protein YPD1. In yeast, YPD1 is normally affected by the cell surface osmosensor histidine kinase SLN1. Under normal conditions, SLN1 is constitutively phosphorylated and its kinase activity maintains YPD1 in its phosphorylated state. YPD1 in turn regulates two parallel pathways by transferring its phosphate to two proteins: SSK1 and SKN7. SSK1 phosphorylation is key to suppressing the yeast’s HOG1 pathway – a stress reponse pathway to high osmolarity, whose activity is lethal under normal conditions. YPD1 also shuttles into the nucleus and activates the SKN7 transcription factor by phosphorylation. In the design, SKN7 was chosen over SSK1 because the SLN1-YPD1-SSK1 pathway is known to affect many high-osmolarity response genes. In order to make SKN7 inducible to IP stimulation (i.e. not constitutively phosphorylated), SLN1 had to be deleted; however, SLN1 deletion is lethal due to activation of the HOG1 pathway. In order to keep the HOG1 pathway silenced in the absence of SLN1, the cells were transformed with a plasmid to overexpress PTP2, an endogenous HOG1 phosphatase, thus rescuing the lethal mutant phenotype.
Using this chassis strain (TM182α) developed by [], the yeast could now be engineered to express the Arabidopsis CRE1 cytokinin receptor. The Weiss group used the CRE1 strain to express IP-inducible GFP in response to sender cells synthesizing IP, as well as a quorum sensing system where the sender and receiver circuits were incorporated into the same cell to achieve cell density dependent expression.
General Mechanism
Project Overview
When large quantities of protein are required for industrial, pharmaceutical and research purposes, researchers may resort to expression of the loci of interest in a genetically modified organism. Once these transgenic organisms are built, they will be grown to sufficient titer before undergoing extraction for the protein of interest. In the case of microorganisms, this is performed in large bioreactors, where the protein of interest will be extracted from a continuously growing cell culture. When the protein of interest is excessively large, synthesis by the organism is difficult, and the use of bioreactors is limited. In the bioreactor, a small subpopulation of cells emerges, where the cells are not expressing the protein correctly. This subpopulation soon dominates due to positive selective pressure, and the use of the bioreactor becomes less effective with time. After a couple of months, the bioreactor will become inefficient, and will have to be “restarted”, a long and costly process. The goal of this year’s uOttawa iGEM team consists of generating a yeast strain capable of long term protein expression.
Pulsate Gene Expression
Gene expression in cells is a dynamic process in which transient responses are often observed. This can be conceptualized as a “pulse” of protein concentration. Achieving such a pulse involves a complex network of activities, which makes it difficult to elucidate the mechanisms in naturally occurring systems. Nevertheless, simple synthetic networks can be created to model this behaviour. We are working on a feed-forward regulatory motif to generate a pulsate expression of a protein of interest, in the model organism Saccharomyces cerevisiae. The general feed-forward mechanism is illustrated in Figure 1A. Upon stimulation (A), a combinatorial promoter is turned on, resulting in expression of the protein of interest (C). The stimulation also turns on another promoter encoding a repressor protein (B), which subsequently acts on the first combinatorial promoter to turn off expression of the protein of interest. Thus, a pulse in protein of interest concentration is achieved.
Cell-to-Cell Communication
An important aspect of naturally occurring biological systems is the cell-to-cell communication that triggers the biomolecular responses in multicellular settings. Our pulse generator system uses signalling elements from Arabidopsis thaliana that have been implemented in yeast [Ref: Chen & Weiss, 2005], to induce the feed-forward network. One population of cells send the signal molecule cytokinin isopentenyladenine (IP), while another population receives the signal via the cytokinin receptor AtCRE1. In combination, the engineered cells receive the IP signal, which induces the pulsate expression of the protein of interest.
Oscillatory Dynamics
After the pulsate protein expression event, the final state of the system consists of a repressed protein of interest. It is desirable to regenerate the system so that it is sensitive to the provided stimulus again – i.e., so that the protein of interest is not repressed. This can be done by destruction of the original inducing signal molecule. Cytokinin dehydrogenase is an enzyme from A. thaliana that degrades the IP signal molecule, and has also been implemented in S. cerevisiae [Ref: Frebortova et al., 2007]. This enzyme is added to the feed-forward network in the manner illustrated in Figure II. Cytokinin dehydrogenase (D) is turned on slowly, and eventually inhibits the stimulus signal (A). With the signal degraded, the repressor protein (B) is no longer produced. With normal protein, any repressor protein present in the cell will inevitably be degraded. Finally, with neither the signal (A) nor the repressor (B) present, the system returns to its initial state.The two populations of sender and receiver cells can be grown together. The inducing IP signal is continually supplied to the receiver cells, which would in turn go through cycles of IP stimulation and degradation. Thus oscillatory dynamics of pulsate protein expression can be accomplished.
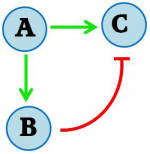
Figure I - General model of the feed forward regulatory motif
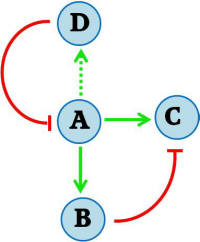
Figure II - An additional element is added to the feed forward regulatory motif, where protein D inhibits the initial stimulus A
Applications
With our pulse generator protein expression system, the proteins are only produced in short pulses, reducing the burden on the cells, and preventing toxic accumulation. These engineered cells can thus be resilient in a bioreactor setting, making them ideal for large-scale protein production applications.
Journal Club
PTP2 Overexpression
- Maeda T, Wurgler-Murphy S M, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242-245
Cytokinin
- Werner T, Motyka V, Strnad M, Schmulling T (2001)Regulation of plant growth by cytokinin. PNAS 98 (18) 10487-10492
Cytokinin Dehydrogenase
- Schmulling T, Werner T, Riefler M, Krupkova E, Bartrina y Manns I (2003) Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res 116:241-252
- Frebortova J, Galuszka P, Werner T, Schmulling T, Frebort I (2007) Functional expression and purification of cytokinin dehydrogenase from Arabidopsis thaliana (AtCKX2) in Saccharomyces cerevisiae. Biologia Plantarium 51 (4):673-682
 "
"