Team:University of Sheffield
From 2008.igem.org
(→On a Mission for Fluorescence...) |
|||
| (139 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | < | + | <!-- If you are creating a new page, start cutting here --> |
| + | [[Image:UniShefBanner.jpg|center]] | ||
| + | {| style="color:#888888;background-color:##888888;" cellpadding="5" cellspacing="2" border="2" bordercolor=#888888 width="85%" align="center" | ||
| + | !align="center"|[[Team:University_of_Sheffield |Introduction]] | ||
| + | !align="center"|[[Team:University_of_Sheffield /Project|Our project]] | ||
| + | !align="center"|[[Team:University_of_Sheffield /Modelling|Modelling]] | ||
| + | !align="center"|[[Team:University_of_Sheffield /Wet Lab|Wet Lab]] | ||
| + | !align="center"|[[Team:University_of_Sheffield /Lab Books| Our team]] | ||
| + | !align="center"|[[Team:University_of_Sheffield /Timetable| Timetable]] | ||
| + | !align="center"|[[Team:University_of_Sheffield /Misc| Miscellaneous]] | ||
| + | |} | ||
| - | < | + | <!-- and finish cutting here! --> |
| + | __NOTOC__ <!-- No table of contents please, this is our homepage. --> | ||
| + | =On a Mission for Fluorescence...= | ||
| + | [[image:GFP_structure.jpg|250px|thumb|right|Structure of GFP courtesy of [http://www.tsienlab.ucsd.edu Tsien Lab]]] | ||
| - | + | Hygiene related diseases have an undeniably huge impact on the world we live in, where many deaths are preventable if sanitary conditions could be improved. This issue is not limited to developing countries: outbursts of diseases caused by water contamination still occur all around the world including the most developed regions. Efficient testing of water is an essential part of preventing these incidents, especially after natural disasters. It needs to be quick, easy enough for anyone to use, cheap and reusable. This was the idea behind the University of Sheffield’s project in its first entry into iGEM. | |
| - | + | By manipulating a pathogen's quorum sensing system and hijacking a pathway already in E.coli, we wanted to build an E.coli cell that could detect ''Vibrio cholerae'' and produce a GFP glow. This would provide the criteria for a good water tester, and hopefully a springboard into the development of other pathogen-detecting E.coli. | |
| + | Adding further depth to the project, our sensor was to be focused around the building of a fusion receptor protein. This would consist of the ‘sensing part’ of the pathogen’s receptor (which detects its own quorum signals) fused to the response regulator of the chosen E.coli receptor. GFP fused downstream of the E.coli receptor’s target genes mean that pathogen present = glow! | ||
| - | + | You can read more about our project [[Team:University_of_Sheffield_/Project | here]], or use the navigation bar above. | |
| - | + | =Sheffield University= | |
| + | [http://www.shef.ac.uk/ The University of Sheffield] is one of the top 'Russel Group' universities in Great Britain, with very highly ranked Molecular Biology and Biotechnology and Engineering departments - which is handy for iGEM! Alternatively, if you fancy a visit, we are home to the best Students Union in the country! | ||
| + | [[Image:Firth_court.png|center|250px|Firth Court, Molecular Biology and Biotechnology department]] | ||
| - | + | =Sponsors= | |
| + | [[Image:ChELSI_Logo_small.png|thumb|left|100x100px|[http://smic.group.shef.ac.uk/04b_ChELSI.html ChELSI]]] | ||
| + | [[Image:Prof_Robert_Poole_logo.png|thumb|left|100x100px|[http://www.shef.ac.uk/mbb/staff/poole/poolelab.html Professor Robert Poole]]] | ||
| + | [[Image:Idtdna-logogif.gif|thumb|left|100x100px|[http://eu.idtdna.com/Home/Home.aspx IDT DNA]]] | ||
| - | + | [[Image:Biofusion_logo.png|thumb|left|100x100px|[http://www.fusionip.co.uk/ Fusion IP]]] | |
| - | + | [[Image:IChemE.jpg|thumb|left|100x100px|[http://www.icheme.org/enetwork/MainFrameset.asp?AreaID=171 Institute of Chemical Engineering]]] | |
| - | + | [[Image:Sheffield_union_logo.PNG|thumb|left|100x100px|[http://www.shef.ac.uk/union/ Sheffield University Union of Students]]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 14:35, 29 October 2008
| Introduction | Our project | Modelling | Wet Lab | Our team | Timetable | Miscellaneous |
|---|
On a Mission for Fluorescence...
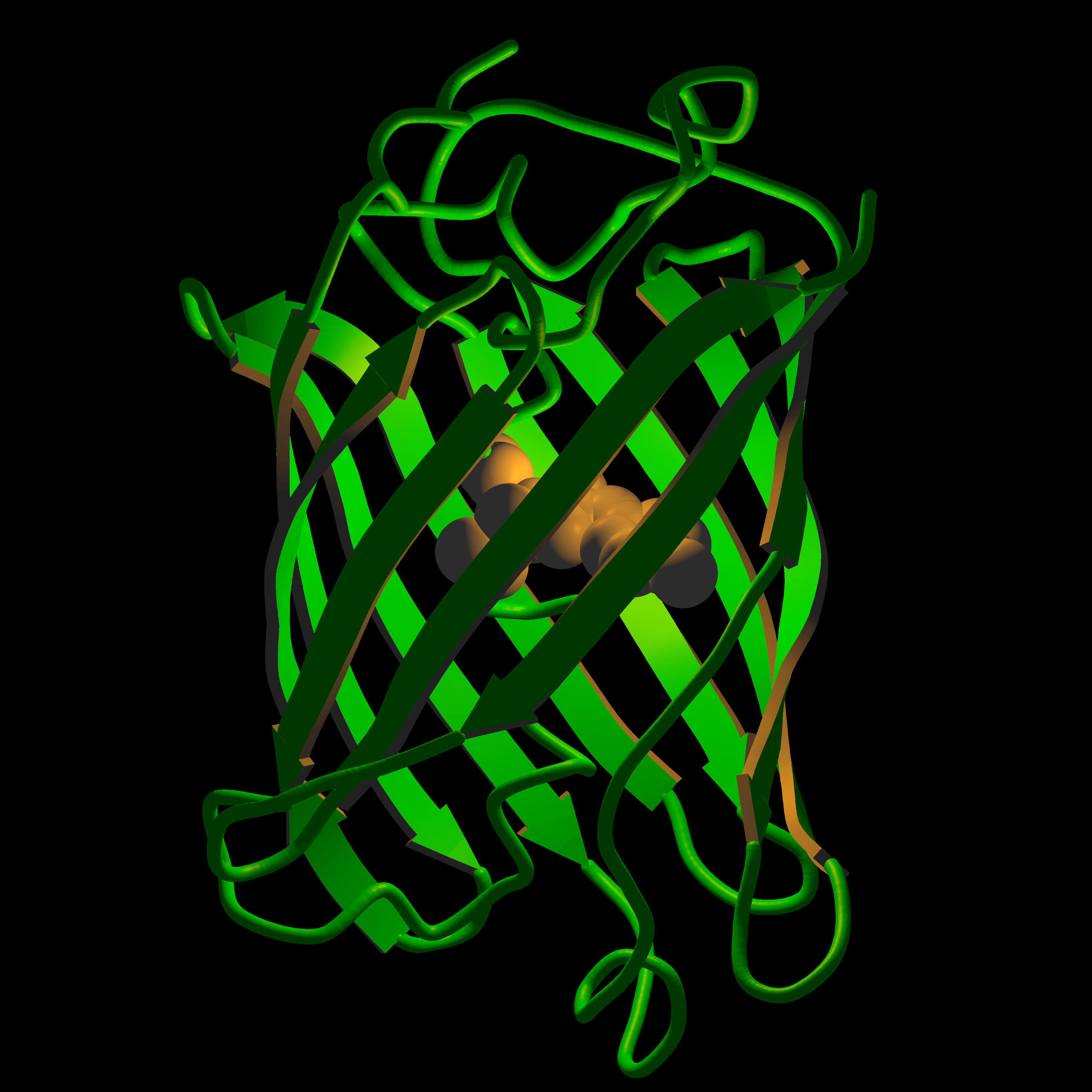
Hygiene related diseases have an undeniably huge impact on the world we live in, where many deaths are preventable if sanitary conditions could be improved. This issue is not limited to developing countries: outbursts of diseases caused by water contamination still occur all around the world including the most developed regions. Efficient testing of water is an essential part of preventing these incidents, especially after natural disasters. It needs to be quick, easy enough for anyone to use, cheap and reusable. This was the idea behind the University of Sheffield’s project in its first entry into iGEM.
By manipulating a pathogen's quorum sensing system and hijacking a pathway already in E.coli, we wanted to build an E.coli cell that could detect Vibrio cholerae and produce a GFP glow. This would provide the criteria for a good water tester, and hopefully a springboard into the development of other pathogen-detecting E.coli. Adding further depth to the project, our sensor was to be focused around the building of a fusion receptor protein. This would consist of the ‘sensing part’ of the pathogen’s receptor (which detects its own quorum signals) fused to the response regulator of the chosen E.coli receptor. GFP fused downstream of the E.coli receptor’s target genes mean that pathogen present = glow!
You can read more about our project here, or use the navigation bar above.
Sheffield University
The University of Sheffield is one of the top 'Russel Group' universities in Great Britain, with very highly ranked Molecular Biology and Biotechnology and Engineering departments - which is handy for iGEM! Alternatively, if you fancy a visit, we are home to the best Students Union in the country!
 "
"



