Team:University of Sheffield /Wet Lab
From 2008.igem.org
(→Lab Books) |
|||
| (31 intermediate revisions not shown) | |||
| Line 12: | Line 12: | ||
!align="center"|[[Team:University_of_Sheffield /Misc| Miscellaneous]] | !align="center"|[[Team:University_of_Sheffield /Misc| Miscellaneous]] | ||
|} | |} | ||
| + | |||
| + | |||
<!-- and finish cutting here! --> | <!-- and finish cutting here! --> | ||
| - | <!-- Nested Wet Lab | + | <!-- Nested Wet Lab menu --> |
| - | {| style="color:#888888;background-color:##888888;" cellpadding=" | + | {| style="color:#888888;background-color:##888888;" cellpadding="1" cellspacing="1" border="4" bordercolor=#888888 width="35%" align="center" |
!align="center"|[[Team:University_of_Sheffield /Wet Lab|Introduction]] | !align="center"|[[Team:University_of_Sheffield /Wet Lab|Introduction]] | ||
!align="center"|[[Team:University_of_Sheffield /Protocols|Protocols]] | !align="center"|[[Team:University_of_Sheffield /Protocols|Protocols]] | ||
|} | |} | ||
| + | |||
=Wet Lab= | =Wet Lab= | ||
| - | + | ||
| - | + | ||
| + | |||
| + | Click [[Team:University_of_Sheffield /Timetable|here]], or on the navigation bar, for the overall timetable of our wet lab sessions. | ||
| + | |||
| + | Click [[Team:University_of_Sheffield /Protocols|here]], or on the sub-navigation bar, for almost all the protocols we used in our wet lab sessions. | ||
| + | |||
| + | The lab books below contain the more detailed jobs carried out by each member of the team. | ||
| + | |||
| + | ==Overview== | ||
| + | |||
| + | [[Image:general_plan.jpg|600px|center|general plan]] | ||
| + | |||
| + | The Diagram is a stepwise representation of a plan we wanted to carry out to achieve our goal. First of all gene coding for BarA protein should be knocked out from wild strain of an engineered bacterium (E.coli MG1655). Then GFP should be introduced into ΔbarA mutant (MGbara1) under a promoter of a gene positively regulated by BarA. Simultaneously to those steps Fusion Kinase should be created and inserted into an expression plasmid. Then both intermediates, ΔbarA mutant with GFP in its genome (MGbara2) and plasmid with Fusion Kinase, should be combined together to give rise to the biosensor (MGfusrec). Functionality of a biosensor should be analyzed by exposing it to CAI-1 autoinducers. If the design is successful the bacterium should grow. | ||
| + | |||
| + | |||
==Lab Books== | ==Lab Books== | ||
<!-- Nasty HTML hack to get the gallery centred --> | <!-- Nasty HTML hack to get the gallery centred --> | ||
<div align=center> | <div align=center> | ||
<gallery align=center perrow="3"> | <gallery align=center perrow="3"> | ||
| - | Image:Dmitry_shef.jpg|[[Team:University_of_Sheffield/Dimtry | | + | Image:Dmitry_shef.jpg|[[Team:University_of_Sheffield/Dimtry | Dmitry's Lab Book]] |
Image:Eva_shef.jpg|[[Team:University_of_Sheffield/Eva | Eva's Lab Book]] | Image:Eva_shef.jpg|[[Team:University_of_Sheffield/Eva | Eva's Lab Book]] | ||
Image:Gosia_shef.jpg|[[Team:University_of_Sheffield/Gosia | Gosia's Lab Book]] | Image:Gosia_shef.jpg|[[Team:University_of_Sheffield/Gosia | Gosia's Lab Book]] | ||
| Line 36: | Line 53: | ||
</div> | </div> | ||
| - | == | + | ==Plasmids and Primers== |
| - | + | {| align="center" border="1" | |
| + | ! colspan="5"|Experimental Primers | ||
| + | |- | ||
| + | |BarA homology | ||
| + | sequences on | ||
| + | pKD13 Kanamycin | ||
| + | resistant cassette | ||
| + | |Forward: | ||
| + | |||
| + | 5' - TGA TGA TTC TGA TCC TGG CAC CGA CCG TCC TTA TTG ATT CCG GGG ATC CGT CGA CC - 3' | ||
| - | = | + | |TM (50mM NaCL): 71.8 °C |
| + | |GC Content: 55.4% | ||
| + | |Molecular Weight (Calculated): 17149.1 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse: | ||
| + | 5' - CGT TGA CTT CGG GCG TCA CGA CGC GAG AGG AAA TAC GTG TAG GCT GGA GCT GCT TC - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 72.6 °C | ||
| + | |GC Content: 58.9% | ||
| + | |Molecular Weight (Calculated): 17377.2 | ||
| + | |- | ||
| + | |RFP insertion | ||
| + | into PGA operon | ||
| + | = PGA homolgy on | ||
| + | RFP cassette | ||
| + | |Forward: | ||
| - | + | 5' - TAA TTA TAC TCA CCA GCA TCA GGA GAT ATT TAT TTC CAT TAC GTA ACA TAT TTA TCC TTA TTA TTA AGC TAC TAA AGC GT - 3' | |
| - | + | |TM (50mM NaCL): 65.6 °C | |
| + | |GC Content: 28.8% | ||
| + | |Molecular Weight (Calculated): 24492 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse: | ||
| + | 5' - AAC TGG CGC GGT TTT GCT GGA TTC GGT TAT GCC GAT GGA CAA TTT AGC GAA GGA AAA GGG ATG GCT TCC TCC GAA GAC GT - 3' | ||
| - | 2. | + | |TM (50mM NaCL): 73.3 °C |
| + | |GC Content: 51.2% | ||
| + | |Molecular Weight (Calculated): 24870.1 | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {| align="center" border="1" | ||
| + | ! colspan="5"|Biobrick Primers | ||
| + | |- | ||
| + | |BIOBRICK 1 + 2 | ||
| + | CsrA with promoter | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''A ACA GAA TGT AAT GCC ATG AC - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 66.6 °C | ||
| + | |GC Content: 48.8% | ||
| + | |Molecular Weight (Calculated): 12618.2 | ||
| + | |- | ||
| + | |CsrA without | ||
| + | promoter | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''A TGC TGA TTC TGA CTC GTC GAG TTG - 3' | ||
| - | + | |TM (50mM NaCL): 68.8 °C | |
| + | |GC Content: 53.3% | ||
| + | |Molecular Weight (Calculated): 13850 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse for both 1 & 2 '''+suffix''': | ||
| + | 5' - '''CTG CAG CGG CCG CTA CTA GTA''' GTA ACT GGA CTG CTG GGA TTT TT - 3' | ||
| - | + | |TM (50mM NaCL): 69 °C | |
| + | |GC Content: 52.3% | ||
| + | |Molecular Weight (Calculated): 13569.8 | ||
| + | |- | ||
| + | |BIOBRICK 3 + 4 | ||
| + | CrsB with | ||
| + | promoter | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''G TCG ACA GGG AGT CAG ACA AC - 3' | ||
| - | + | |TM (50mM NaCL): 69.9 °C | |
| + | |GC Content: 58.5% | ||
| + | |Molecular Weight (Calculated): 12660.2 | ||
| + | |- | ||
| + | |CsrB without | ||
| + | promoter | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''A GGG AGT CAG ACA ACG AAG T - 3' | ||
| - | + | |TM (50mM NaCL): 69 °C | |
| + | |GC Content: 55% | ||
| + | |Molecular Weight (Calculated): 12395.1 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse for both 3 & 4 '''+suffix''': | ||
| + | 5' - '''CTG CAG CGG CCG CTA CTA GTA''' AAT AAA AAA AGG GAG CAC TG - 3' | ||
| - | + | |TM (50mM NaCL): 66.5 °C | |
| + | |GC Content: 48.8% | ||
| + | |Molecular Weight (Calculated): 12685.3 | ||
| + | |- | ||
| + | |BIOBRICKS 5 & 6 | ||
| + | UvrY with | ||
| + | promoter | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''A ATG ACT AAC TAT CAG TAG CGT TAT C - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 65.5 °C | ||
| + | |GC Content: 45.7% | ||
| + | |Molecular Weight (Calculated): 14124.2 | ||
| + | |- | ||
| + | |UvrY without | ||
| + | promoter | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''T ATT CCT TTG ATC AAC GTT CTA C - 3' | ||
| - | ''' | + | |TM (50mM NaCL): 65.7 °C |
| + | |GC Content: 46.5% | ||
| + | |Molecular Weight (Calculated): 13110.5 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse for both 5 & 6 '''+suffix''' | ||
| + | 5' - '''CTG CAG CGG CCG CTA CTA GTA''' TCA CTG ACT TGA TAA TGT CT - 3' | ||
| - | + | |TM (50mM NaCL): 66.3 °C | |
| - | + | |GC Content: 48.8% | |
| + | |Molecular Weight (Calculated): 12551.2 | ||
| + | |- | ||
| + | |BIOBRICK 7 | ||
| + | BarA | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''C GGA ACT CCA TGA CCA ACT ACA G - 3' | ||
| - | + | |TM (50mM NaCL): 69.6 °C | |
| - | + | |GC Content: 55.8% | |
| - | + | |Molecular Weight (Calculated): 13157.6 | |
| - | + | |- | |
| - | + | | | |
| - | + | |Reverse '''+suffix''': | |
| - | + | 5' - '''CTG CAG CGG CCG CTA CTA GTA''' TTA CCC GAG AAT TTT GCT GG - 3' | |
| - | + | ||
| - | + | ||
| - | ''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ''' | + | |
| - | + | ||
| - | + | ||
| - | + | |TM (50mM NaCL): 68.2 °C | |
| + | |GC Content: 53.7% | ||
| + | |Molecular Weight (Calculated): 12592.2 | ||
| + | |- | ||
| + | |BIOBRICK 8 | ||
| + | Ori101 of pKD46 | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''A GTC TCA ATT GGT CTA GGT GAT - 3' | ||
| - | ''' | + | |TM (50mM NaCL): 67.3 °C |
| + | |GC Content: 50% | ||
| + | |Molecular Weight (Calculated): 12951.4 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse '''+suffix''': | ||
| + | 5' - '''CTG CAG CGG CCG CTA CTA GTA''' GTT GAT GAT ACC GCT GCC TT - 3' | ||
| - | + | |TM (50mM NaCL): 69.8 °C | |
| - | + | |GC Content: 56.1% | |
| - | + | |Molecular Weight (Calculated): 12568.2 | |
| - | + | |- | |
| + | |BIOBRICK 9 | ||
| + | CqsA - the CAI-1 | ||
| + | producer | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''A TGA ACA AAC CGC AGC TGC C - 3' | ||
| - | ''' | + | |TM (50mM NaCL): 70.4 °C |
| + | |GC Content: 57.5% | ||
| + | |Molecular Weight (Calculated): 12251 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse '''+suffix''' | ||
| + | 5' - '''CTG CAG CGG CCG CTA CTA GTA''' ACG GAA GTA GAA GTC ACC GT - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 69.3 °C | ||
| + | |GC Content: 56.1% | ||
| + | |Molecular Weight (Calculated): 12644.2 | ||
| + | |- | ||
| + | |BIOBRICK 10 | ||
| + | CqsS - the gene | ||
| + | we synthesised: | ||
| + | CAI-1 receptor | ||
| + | |Forward '''+prefix''' | ||
| + | 5' - GAA TTC GCG GCC GCT TCT AG'''A TGA TCG TTT CTA TGG ACG T - 3' | ||
| - | + | |TM (50mM NaCL): 67 °C | |
| - | + | |GC Content: 50% | |
| - | + | |Molecular Weight (Calculated): 12309 | |
| - | + | |- | |
| - | + | | | |
| - | + | |Reverse '''+suffix''' | |
| + | 5' - CTG CAG CGG CCG CTA CTA GTA''' TTA AAC CCA CGC CGC AAC TT - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 70.4 °C | ||
| + | |GC Content: 56.1% | ||
| + | |Molecular Weight (Calculated): 12475.1 | ||
| + | |- | ||
| + | |BIOBRICK 11 | ||
| + | Red recombinase | ||
| + | system | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - GAA TTC GCG GCC GCT TCT AG'''A TGG ATA TTA ATA CTG AAA C - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 63.1 °C | ||
| + | |GC Content: 42.5% | ||
| + | |Molecular Weight (Calculated): 12319 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse '''+suffix''': | ||
| + | 5' - '''CTG CAG CGG CCG CTA CTA GTA''' TCA TCG CCA TTG CTC CCC AA - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 71.2 °C | ||
| + | |GC Content: 58.5% | ||
| + | |Molecular Weight (Calculated): 12442.1 | ||
| + | |- | ||
| + | |BIOBRICK 12 | ||
| + | FLP recombinase | ||
| + | |Forward '''+prefix''': | ||
| + | 5' - '''GAA TTC GCG GCC GCT TCT AG'''A TGT CTA GTT ACA TGG ATT T - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 64.9 °C | ||
| + | |GC Content: 45% | ||
| + | |Molecular Weight (Calculated): 12308 | ||
| + | |- | ||
| + | | | ||
| + | |Reverse '''+suffix''': | ||
| + | 5' - '''CTG CAG CGG CCG CTA CTA GTA''' GGT ACC TTT TTA GAA AAA TT - 3' | ||
| + | |||
| + | |TM (50mM NaCL): 64.9 °C | ||
| + | |GC Content: 43.9% | ||
| + | |Molecular Weight (Calculated): 12599.2 | ||
| + | |} | ||
| - | + | Plasmids: | |
| - | * | + | [[Image:pKD46.png]] |
| - | * | + | [[Image:pKD13.png]] |
| + | *pCP20 - "an AmpR and CmR plasmid that shows temperature-sensitive replication and thermal induction of FLP synthesis" (Datsenko and Wanner,[4]) | ||
| + | *pCQSA - plasmid sent to us by Bonnie Bassler's lab, contains the CAI-1 producing gene | ||
| + | *pCQSS - plasmid synthesised by us (via idtDNA) containing the gene for the CAI-1 receptor | ||
Latest revision as of 01:49, 30 October 2008
| Introduction | Our project | Modelling | Wet Lab | Our team | Timetable | Miscellaneous |
|---|
| Introduction | Protocols |
|---|
Contents |
Wet Lab
Click here, or on the navigation bar, for the overall timetable of our wet lab sessions.
Click here, or on the sub-navigation bar, for almost all the protocols we used in our wet lab sessions.
The lab books below contain the more detailed jobs carried out by each member of the team.
Overview
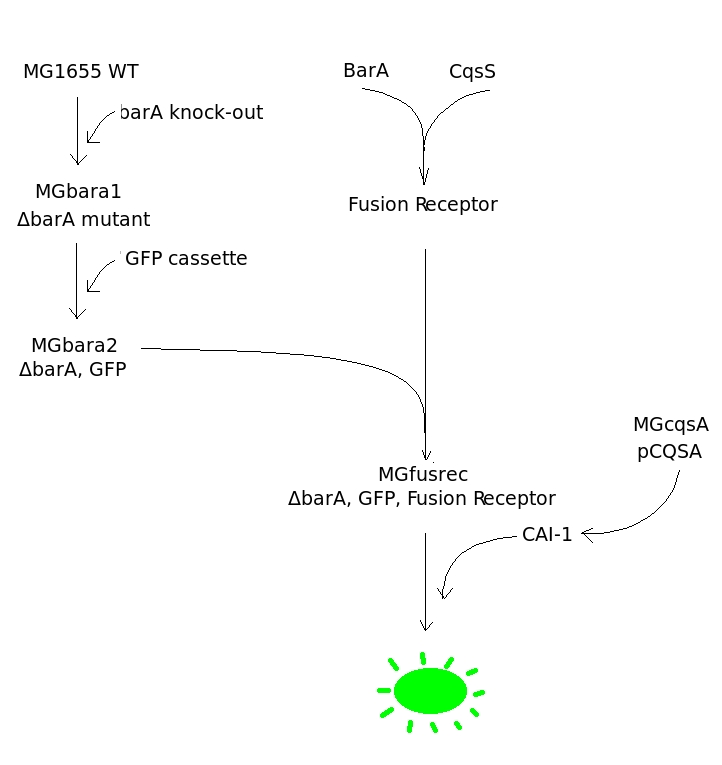
The Diagram is a stepwise representation of a plan we wanted to carry out to achieve our goal. First of all gene coding for BarA protein should be knocked out from wild strain of an engineered bacterium (E.coli MG1655). Then GFP should be introduced into ΔbarA mutant (MGbara1) under a promoter of a gene positively regulated by BarA. Simultaneously to those steps Fusion Kinase should be created and inserted into an expression plasmid. Then both intermediates, ΔbarA mutant with GFP in its genome (MGbara2) and plasmid with Fusion Kinase, should be combined together to give rise to the biosensor (MGfusrec). Functionality of a biosensor should be analyzed by exposing it to CAI-1 autoinducers. If the design is successful the bacterium should grow.
Lab Books
Plasmids and Primers
| Experimental Primers | ||||
|---|---|---|---|---|
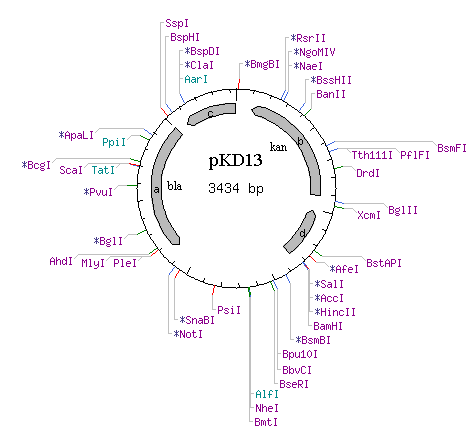
| BarA homology
sequences on pKD13 Kanamycin resistant cassette | Forward:
5' - TGA TGA TTC TGA TCC TGG CAC CGA CCG TCC TTA TTG ATT CCG GGG ATC CGT CGA CC - 3' | TM (50mM NaCL): 71.8 °C | GC Content: 55.4% | Molecular Weight (Calculated): 17149.1 |
| Reverse:
5' - CGT TGA CTT CGG GCG TCA CGA CGC GAG AGG AAA TAC GTG TAG GCT GGA GCT GCT TC - 3' | TM (50mM NaCL): 72.6 °C | GC Content: 58.9% | Molecular Weight (Calculated): 17377.2 | |
| RFP insertion
into PGA operon = PGA homolgy on RFP cassette | Forward:
5' - TAA TTA TAC TCA CCA GCA TCA GGA GAT ATT TAT TTC CAT TAC GTA ACA TAT TTA TCC TTA TTA TTA AGC TAC TAA AGC GT - 3' | TM (50mM NaCL): 65.6 °C | GC Content: 28.8% | Molecular Weight (Calculated): 24492 |
| Reverse:
5' - AAC TGG CGC GGT TTT GCT GGA TTC GGT TAT GCC GAT GGA CAA TTT AGC GAA GGA AAA GGG ATG GCT TCC TCC GAA GAC GT - 3' | TM (50mM NaCL): 73.3 °C | GC Content: 51.2% | Molecular Weight (Calculated): 24870.1 | |
| Biobrick Primers | ||||
|---|---|---|---|---|
| BIOBRICK 1 + 2
CsrA with promoter | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGA ACA GAA TGT AAT GCC ATG AC - 3' | TM (50mM NaCL): 66.6 °C | GC Content: 48.8% | Molecular Weight (Calculated): 12618.2 |
| CsrA without
promoter | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGA TGC TGA TTC TGA CTC GTC GAG TTG - 3' | TM (50mM NaCL): 68.8 °C | GC Content: 53.3% | Molecular Weight (Calculated): 13850 |
| Reverse for both 1 & 2 +suffix:
5' - CTG CAG CGG CCG CTA CTA GTA GTA ACT GGA CTG CTG GGA TTT TT - 3' | TM (50mM NaCL): 69 °C | GC Content: 52.3% | Molecular Weight (Calculated): 13569.8 | |
| BIOBRICK 3 + 4
CrsB with promoter | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGG TCG ACA GGG AGT CAG ACA AC - 3' | TM (50mM NaCL): 69.9 °C | GC Content: 58.5% | Molecular Weight (Calculated): 12660.2 |
| CsrB without
promoter | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGA GGG AGT CAG ACA ACG AAG T - 3' | TM (50mM NaCL): 69 °C | GC Content: 55% | Molecular Weight (Calculated): 12395.1 |
| Reverse for both 3 & 4 +suffix:
5' - CTG CAG CGG CCG CTA CTA GTA AAT AAA AAA AGG GAG CAC TG - 3' | TM (50mM NaCL): 66.5 °C | GC Content: 48.8% | Molecular Weight (Calculated): 12685.3 | |
| BIOBRICKS 5 & 6
UvrY with promoter | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGA ATG ACT AAC TAT CAG TAG CGT TAT C - 3' | TM (50mM NaCL): 65.5 °C | GC Content: 45.7% | Molecular Weight (Calculated): 14124.2 |
| UvrY without
promoter | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGT ATT CCT TTG ATC AAC GTT CTA C - 3' | TM (50mM NaCL): 65.7 °C | GC Content: 46.5% | Molecular Weight (Calculated): 13110.5 |
| Reverse for both 5 & 6 +suffix
5' - CTG CAG CGG CCG CTA CTA GTA TCA CTG ACT TGA TAA TGT CT - 3' | TM (50mM NaCL): 66.3 °C | GC Content: 48.8% | Molecular Weight (Calculated): 12551.2 | |
| BIOBRICK 7
BarA | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGC GGA ACT CCA TGA CCA ACT ACA G - 3' | TM (50mM NaCL): 69.6 °C | GC Content: 55.8% | Molecular Weight (Calculated): 13157.6 |
| Reverse +suffix:
5' - CTG CAG CGG CCG CTA CTA GTA TTA CCC GAG AAT TTT GCT GG - 3' | TM (50mM NaCL): 68.2 °C | GC Content: 53.7% | Molecular Weight (Calculated): 12592.2 | |
| BIOBRICK 8
Ori101 of pKD46 | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGA GTC TCA ATT GGT CTA GGT GAT - 3' | TM (50mM NaCL): 67.3 °C | GC Content: 50% | Molecular Weight (Calculated): 12951.4 |
| Reverse +suffix:
5' - CTG CAG CGG CCG CTA CTA GTA GTT GAT GAT ACC GCT GCC TT - 3' | TM (50mM NaCL): 69.8 °C | GC Content: 56.1% | Molecular Weight (Calculated): 12568.2 | |
| BIOBRICK 9
CqsA - the CAI-1 producer | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGA TGA ACA AAC CGC AGC TGC C - 3' | TM (50mM NaCL): 70.4 °C | GC Content: 57.5% | Molecular Weight (Calculated): 12251 |
| Reverse +suffix
5' - CTG CAG CGG CCG CTA CTA GTA ACG GAA GTA GAA GTC ACC GT - 3' | TM (50mM NaCL): 69.3 °C | GC Content: 56.1% | Molecular Weight (Calculated): 12644.2 | |
| BIOBRICK 10
CqsS - the gene we synthesised: CAI-1 receptor | Forward +prefix
5' - GAA TTC GCG GCC GCT TCT AGA TGA TCG TTT CTA TGG ACG T - 3' | TM (50mM NaCL): 67 °C | GC Content: 50% | Molecular Weight (Calculated): 12309 |
| Reverse +suffix
5' - CTG CAG CGG CCG CTA CTA GTA TTA AAC CCA CGC CGC AAC TT - 3' | TM (50mM NaCL): 70.4 °C | GC Content: 56.1% | Molecular Weight (Calculated): 12475.1 | |
| BIOBRICK 11
Red recombinase system | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGA TGG ATA TTA ATA CTG AAA C - 3' | TM (50mM NaCL): 63.1 °C | GC Content: 42.5% | Molecular Weight (Calculated): 12319 |
| Reverse +suffix:
5' - CTG CAG CGG CCG CTA CTA GTA TCA TCG CCA TTG CTC CCC AA - 3' | TM (50mM NaCL): 71.2 °C | GC Content: 58.5% | Molecular Weight (Calculated): 12442.1 | |
| BIOBRICK 12
FLP recombinase | Forward +prefix:
5' - GAA TTC GCG GCC GCT TCT AGA TGT CTA GTT ACA TGG ATT T - 3' | TM (50mM NaCL): 64.9 °C | GC Content: 45% | Molecular Weight (Calculated): 12308 |
| Reverse +suffix:
5' - CTG CAG CGG CCG CTA CTA GTA GGT ACC TTT TTA GAA AAA TT - 3' | TM (50mM NaCL): 64.9 °C | GC Content: 43.9% | Molecular Weight (Calculated): 12599.2 | |
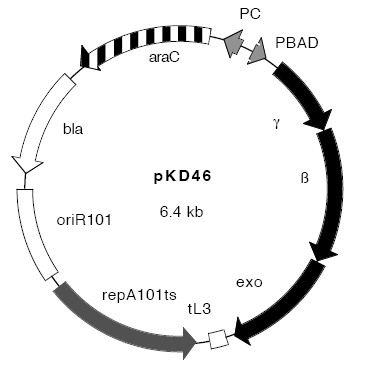
Plasmids:
- pCP20 - "an AmpR and CmR plasmid that shows temperature-sensitive replication and thermal induction of FLP synthesis" (Datsenko and Wanner,[4])
- pCQSA - plasmid sent to us by Bonnie Bassler's lab, contains the CAI-1 producing gene
- pCQSS - plasmid synthesised by us (via idtDNA) containing the gene for the CAI-1 receptor
 "
"